The 4EHP-mediated translational repression of cGAS impedes the host immune response against DNA viruses
- PMID: 39560640
- PMCID: PMC11621783
- DOI: 10.1073/pnas.2413018121
The 4EHP-mediated translational repression of cGAS impedes the host immune response against DNA viruses
Abstract
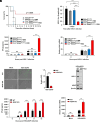
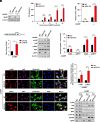
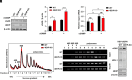
A critical host response against viral infections entails the activation of innate immune signaling that culminates in the production of antiviral proteins. DNA viruses are sensed by the cytosolic pattern recognition receptor cyclic GMP-AMP synthase (cGAS), which initiates a signaling pathway that results in production of proinflammatory cytokines such as Interferon-β (IFN-β) and activation of the antiviral response. Precise regulation of the antiviral innate immune response is required to avoid deleterious effects of its overactivation. We previously reported that the 4EHP/GIGYF2 translational repressor complex reduces the translation of Ifnb1 mRNA, which encodes IFN-β, upon RNA viral infections. Here, we report a distinct regulatory mechanism by which 4EHP controls replication of DNA viruses by translational repression of the Cgas mRNA, which encodes the DNA viral sensor cGAS. We show that 4EHP is required for effective translational repression of Cgas mRNA triggered by miR-23a. Upon infection, 4EHP deficiency bolsters the elicited innate immune response against the diverse DNA viruses Herpes simplex virus 1 (HSV-1) and Vaccinia Virus (VacV) and concomitantly reduces their rate of replication in vitro and in vivo. This study elucidates an intrinsic regulatory mechanism of the host response to DNA viruses which may provide unique opportunities for countering viral infections.
Keywords: 4EHP; DNA virus; cGAS; innate immunity; microRNA.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Seth R. B., Sun L., Ea C. K., Chen Z. J., Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669–682 (2005). - PubMed
MeSH terms
Substances
Grants and funding
- BB/W008165/1/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- R259964C0G/Canadian Government | Canadian Institutes of Health Research (CIHR)
- 2020R1A6A3A03040141/National Research Foundation of Korea (NRF)
- FDN-148423/Canadian Government | Canadian Institutes of Health Research (CIHR)
- 1073/Terry Fox Research Institute (TFRI)
LinkOut - more resources
Full Text Sources
Miscellaneous

