Selective regulation of IFN-γ and IL-4 co-producing unconventional T cells by purinergic signaling
- PMID: 39560665
- PMCID: PMC11577439
- DOI: 10.1084/jem.20240354
Selective regulation of IFN-γ and IL-4 co-producing unconventional T cells by purinergic signaling
Abstract
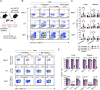
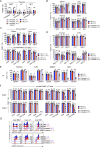
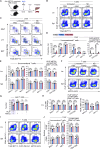
Unconventional T cells, including mucosal-associated invariant T (MAIT), natural killer T (NKT), and gamma-delta T (γδT) cells, comprise distinct T-bet+, IFN-γ+ and RORγt+, IL-17+ subsets which play differential roles in health and disease. NKT1 cells are susceptible to ARTC2-mediated P2X7 receptor (P2RX7) activation, but the effects on other unconventional T-cell types are unknown. Here, we show that MAIT, γδT, and NKT cells express P2RX7 and are sensitive to P2RX7-mediated cell death. Mouse peripheral T-bet+ MAIT1, γδT1, and NKT1 cells, especially in liver, co-express ARTC2 and P2RX7. These markers could be further upregulated upon exposure to retinoic acid. Blocking ARTC2 or inhibiting P2RX7 protected MAIT1, γδT1, and NKT1 cells from cell death, enhanced their survival in vivo, and increased the number of IFN-γ-secreting cells without affecting IL-17 production. Importantly, this revealed the existence of IFN-γ and IL-4 co-producing unconventional T-cell populations normally lost upon isolation due to ARTC2/P2RX7-induced death. Administering extracellular NAD in vivo activated this pathway, depleting P2RX7-sensitive unconventional T cells. Our study reveals ARTC2/P2RX7 as a common regulatory axis modulating the unconventional T-cell compartment, affecting the viability of IFN-γ- and IL-4-producing T cells, offering important insights to facilitate future studies into how these cells can be regulated in health and disease.
© 2024 Xu et al.
Conflict of interest statement
Disclosures: A. Brandli reported personal fees from F. Hoffmann-La Roche Ltd outside the submitted work. J.Y.W. Mak reported a patent to WO2015149130A9 licensed. D.I. Godfrey reported personal fees from Avalia Immunotherapies outside the submitted work; in addition, D.I. Godfrey had a patent to PCT/AU2020/050662 pending and a patent to PCT/WO2020/231274 pending. No other disclosures were reported.
Figures










References
-
- Ataide, M.A., Knopper K., Cruz de Casas P., Ugur M., Eickhoff S., Zou M., Shaikh H., Trivedi A., Grafen A., Yang T., et al. . 2022. Lymphatic migration of unconventional T cells promotes site-specific immunity in distinct lymph nodes. Immunity. 55:1813–1828.e9. 10.1016/j.immuni.2022.07.019 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

