Immunomolecular and reactivity landscapes of gut IgA subclasses in homeostasis and inflammatory bowel disease
- PMID: 39560666
- PMCID: PMC11577441
- DOI: 10.1084/jem.20230079
Immunomolecular and reactivity landscapes of gut IgA subclasses in homeostasis and inflammatory bowel disease
Abstract
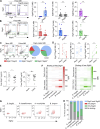
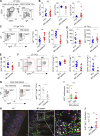
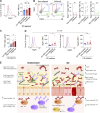
The human gut includes plasma cells (PCs) expressing immunoglobulin A1 (IgA1) or IgA2, two structurally distinct IgA subclasses with elusive regulation, function, and reactivity. We show here that intestinal IgA1+ and IgA2+ PCs co-emerged early in life, comparably accumulated somatic mutations, and were enriched within short-lived CD19+ and long-lived CD19- PC subsets, respectively. IgA2+ PCs were extensively clonally related to IgA1+ PCs and a subset of them presumably emerged from IgA1+ precursors. Of note, secretory IgA1 (SIgA1) and SIgA2 dually coated a large fraction of mucus-embedded bacteria, including Akkermansia muciniphila. Disruption of homeostasis by inflammatory bowel disease (IBD) was associated with an increase in actively proliferating IgA1+ plasmablasts, a depletion in long-lived IgA2+ PCs, and increased SIgA1+SIgA2+ gut microbiota. Such increase featured enhanced IgA1 reactivity to pathobionts, including Escherichia coli, combined with depletion of beneficial A. muciniphila. Thus, gut IgA1 and IgA2 emerge from clonally related PCs and show unique changes in both frequency and reactivity in IBD.
© 2024 Tejedor Vaquero et al.
Conflict of interest statement
Disclosures: L. Comerma reported personal fees from Roche, MSD, AstraZeneca, and Diaceutics and non-financial support from Roche, MSD, AstraZeneca, and Phillips outside the submitted work. M. Iglesias reported personal fees from Bristol Myers Squibb, Merck Sharp & Dohme, Roche, Astellas, Merck, Agilent, and Seagen outside the submitted work. No other disclosures were reported.
Figures







References
-
- Ansaldo, E., Slayden L.C., Ching K.L., Koch M.A., Wolf N.K., Plichta D.R., Brown E.M., Graham D.B., Xavier R.J., Moon J.J., and Barton G.M.. 2019. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 364:1179–1184. 10.1126/science.aaw7479 - DOI - PMC - PubMed
-
- Berkowska, M.A., Driessen G.J.A., Bikos V., Grosserichter-Wagener C., Stamatopoulos K., Cerutti A., He B., Biermann K., Lange J.F., van der Burg M., et al. . 2011. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood. 118:2150–2158. 10.1182/blood-2011-04-345579 - DOI - PMC - PubMed
-
- Bolyen, E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. . 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:852–857. 10.1038/s41587-019-0209-9 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

