G-CSF resistance of ELANE-mutant neutropenia depends on SERF1-containing truncated-neutrophil elastase aggregates
- PMID: 39560992
- PMCID: PMC11735094
- DOI: 10.1172/JCI177342
G-CSF resistance of ELANE-mutant neutropenia depends on SERF1-containing truncated-neutrophil elastase aggregates
Abstract
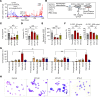
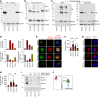
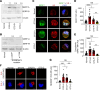
Severe congenital neutropenia (SCN) is frequently associated with dominant point mutations in ELANE, the gene encoding neutrophil elastase (NE). Chronic administration of granulocyte colony-stimulating factor (G-CSF) is a first-line treatment of ELANE-mutant (ELANEmut) SCN. However, some ELANEmut patients, including patients with ELANE start codon mutations, do not respond to G-CSF. Here, through directed granulopoiesis of gene-edited isogenic normal and patient-derived iPSCs, we demonstrate that ELANE start codon mutations suffice to induce G-CSF-resistant granulocytic precursor cell death and refractory SCN. ELANE start codon-mutated neutrophil precursors express predominantly nuclear N-terminally truncated alternate NE. Unlike G-CSF-sensitive ELANE mutations that induce endoplasmic reticulum and unfolded protein response stress, we found that the mutation of the ELANE translation initiation codon resulted in NE aggregates and activated proapoptotic aggrephagy, as determined by downregulated BAG1 expression, decreased BAG1/BAG3 ratio, NE colocalization with BAG3, and localized expression of autophagic LC3B. We found that SERF1, an RNA-chaperone protein, known to localize in misfolded protein aggregates in neurodegenerative diseases, was highly upregulated and interacted with cytoplasmic NE of mutant neutrophil precursors. Silencing of SERF1 enhanced survival and differentiation of iPSC-derived neutrophil precursors, restoring their responsiveness to G-CSF. These observations provide a mechanistic insight into G-CSF-resistant ELANEmut SCN, revealing targets for therapeutic intervention.
Keywords: Hematology; Neutrophils.
Conflict of interest statement
Figures

References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

