Low tristetraprolin expression activates phenotypic plasticity and primes transition to lethal prostate cancer in mice
- PMID: 39560993
- PMCID: PMC11735106
- DOI: 10.1172/JCI175680
Low tristetraprolin expression activates phenotypic plasticity and primes transition to lethal prostate cancer in mice
Abstract
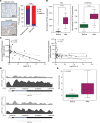
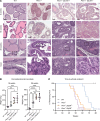
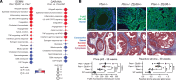
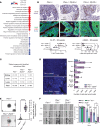
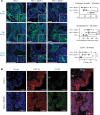
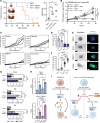
Phenotypic plasticity is a hallmark of cancer and is increasingly realized as a mechanism of resistance to androgen receptor-targeted (AR-targeted) therapy. Now that many prostate cancer (PCa) patients are treated upfront with AR-targeted agents, it is critical to identify actionable mechanisms that drive phenotypic plasticity, to prevent the emergence of resistance. We showed that loss of tristetraprolin (TTP; gene ZFP36) increased NF-κB activation, and was associated with higher rates of aggressive disease and early recurrence in primary PCa. We also examined the clinical and biological impact of ZFP36 loss with co-loss of PTEN, a known driver of PCa. Analysis of multiple independent primary PCa cohorts demonstrated that PTEN and ZFP36 co-loss was associated with increased recurrence risk. Engineering prostate-specific Zfp36 deletion in vivo induced prostatic intraepithelial neoplasia, and, with Pten codeletion, resulted in rapid progression to castration-resistant adenocarcinoma. Zfp36 loss altered the cell state driven by Pten loss, as demonstrated by enrichment of epithelial-mesenchymal transition (EMT), inflammation, TNF-α/NF-κB, and IL-6-JAK/STAT3 gene sets. Additionally, our work revealed that ZFP36 loss also induced enrichment of multiple gene sets involved in mononuclear cell migration, chemotaxis, and proliferation. Use of the NF-κB inhibitor dimethylaminoparthenolide (DMAPT) induced marked therapeutic responses in tumors with PTEN and ZFP36 co-loss and reversed castration resistance.
Keywords: Cell biology; Mouse models; Oncology; Prostate cancer.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

