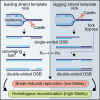
DNA nicks in both leading and lagging strand templates can trigger break-induced replication
- PMID: 39561776
- PMCID: PMC12095120
- DOI: 10.1016/j.molcel.2024.10.026
DNA nicks in both leading and lagging strand templates can trigger break-induced replication
Abstract
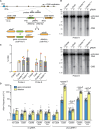
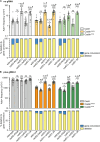
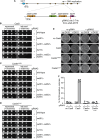
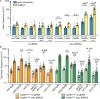
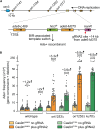
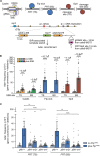
Encounters between replication forks and unrepaired DNA single-strand breaks (SSBs) can generate both single-ended and double-ended double-strand breaks (seDSBs and deDSBs). seDSBs can be repaired by break-induced replication (BIR), which is a highly mutagenic pathway that is thought to be responsible for many of the mutations and genome rearrangements that drive cancer development. However, the frequency of BIR's deployment and its ability to be triggered by both leading and lagging template strand SSBs were unclear. Using site- and strand-specific SSBs generated by nicking enzymes, including CRISPR-Cas9 nickase (Cas9n), we demonstrate that leading and lagging template strand SSBs in fission yeast are typically converted into deDSBs that are repaired by homologous recombination. However, both types of SSBs can also trigger BIR, and the frequency of these events increases when fork convergence is delayed and the non-homologous end joining protein Ku70 is deleted.
Keywords: CRISPR-Cas9n; DNA replication; Flp-nick; break-induced replication; double-strand break; homologous recombination; replication fork; replication fork collapse; replication restart; single-strand break.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

