Why HPV16? Why, now, HPV42? How the discovery of HPV42 in rare cancers provides an opportunity to challenge our understanding about the transition between health and disease for common members of the healthy microbiota
- PMID: 39562287
- PMCID: PMC11644485
- DOI: 10.1093/femsre/fuae029
Why HPV16? Why, now, HPV42? How the discovery of HPV42 in rare cancers provides an opportunity to challenge our understanding about the transition between health and disease for common members of the healthy microbiota
Abstract
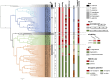
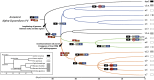
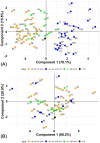
In 2022, a bioinformatic, agnostic approach identified HPV42 as causative agent of a rare cancer, later confirmed experimentally. This unexpected association offers an opportunity to reconsider our understanding about papillomavirus infections and cancers. We have expanded our knowledge about the diversity of papillomaviruses and the diseases they cause. Yet, we still lack answers to fundamental questions, such as what makes HPV16 different from the closely related HPV31 or HPV33; or why the very divergent HPV13 and HPV32 cause focal epithelial hyperplasia, while HPV6 or HPV42 do not, despite their evolutionary relatedness. Certain members of the healthy skin microbiota are associated to rare clinical conditions. We propose that a focus on cellular phenotypes, most often transient and influenced by intrinsic and extrinsic factors, may help understand the continuum between health and disease. A conceptual switch is required towards an interpretation of biology as a diversity of states connected by transition probabilities, rather than quasi-deterministic programs. Under this perspective, papillomaviruses may only trigger malignant transformation when specific viral genotypes interact with precise cellular states. Drawing on Canguilhem's concepts of normal and pathological, we suggest that understanding the transition between fluid cellular states can illuminate how commensal-like infections transition from benign to malignant.
Keywords: cell type; diversity; ecology; evolution; genotype-to-phenotype; infection and cancer; microbiote; papillomavirus; phenotype; phenotype-to-environment; virome; virus–host interactions.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures
References
-
- Arbyn M, Tommasino M, Depuydt C et al. Are 20 human papillomavirus types causing cervical cancer?. J Pathol. 2014;234:431–5. - PubMed
-
- Archard HO, Heck JW, Stanley HR. Focal epithelial hyperplasia: an unusual oral mucosal lesion found in indian children. Oral Surg Oral Med Oral Pathol. 1965;20:201–12. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

