Linking the transcriptome to physiology: response of the proteome of Cupriavidus metallidurans to changing metal availability
- PMID: 39562290
- PMCID: PMC11647595
- DOI: 10.1093/mtomcs/mfae058
Linking the transcriptome to physiology: response of the proteome of Cupriavidus metallidurans to changing metal availability
Abstract
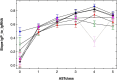
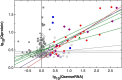
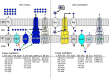
Cupriavidus metallidurans CH34 is a metal-resistant bacterium. Its metal homeostasis is based on a flow equilibrium of metal ion uptake and efflux reactions, which adapts to changing metal concentrations within an hour. At high metal concentrations, upregulation of the genes for metal efflux systems occurs within minutes. Here, we investigate the changes in the bacterial proteome accompanying these genetic and physiological events after 1.5 cell duplications, which took 3 h. To that end, C. metallidurans CH34 and its plasmid-free derivative, AE104, either were challenged with a toxic metal mix or were cultivated under metal-starvation conditions, followed by bottom-up proteomics. When metal-shocked or -starved cells were compared with their respective controls, 3540 proteins changed in abundance, with 76% appearing in one, but not the other, condition; the remaining 24% were up- or downregulated. Metal-shocked C. metallidurans strains had adjusted their proteomes to combat metal stress. The most prominent polypeptides were the products of the plasmid-encoded metal-resistance determinants in strain CH34, particularly the CzcCBA transenvelope efflux system. Moreover, the influence of antisense transcripts on the proteome was also revealed. In one specific example, the impact of an asRNA on the abundance of gene products could be demonstrated and this yielded new insights into the function of the transmembrane efflux complex ZniCBA under conditions of metal starvation.
Keywords: Cupriavidus metallidurans; metal homeostasis; metal starvation; proteomics; transenvelope efflux systems; zinc.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Antisense transcription is associated with expression of metal resistance determinants in Cupriavidus metallidurans CH34.Metallomics. 2024 Dec 2;16(12):mfae057. doi: 10.1093/mtomcs/mfae057. Metallomics. 2024. PMID: 39562278 Free PMC article.
-
Deletion of the zupT gene for a zinc importer influences zinc pools in Cupriavidus metallidurans CH34.Metallomics. 2014 Mar;6(3):421-36. doi: 10.1039/c3mt00267e. Epub 2014 Jan 10. Metallomics. 2014. PMID: 24407051
-
Loss of Mobile Genomic Islands in Metal-Resistant, Hydrogen-Oxidizing Cupriavidus metallidurans.Appl Environ Microbiol. 2022 Feb 22;88(4):e0204821. doi: 10.1128/AEM.02048-21. Epub 2021 Dec 15. Appl Environ Microbiol. 2022. PMID: 34910578 Free PMC article.
-
Cupriavidus metallidurans: evolution of a metal-resistant bacterium.Antonie Van Leeuwenhoek. 2009 Aug;96(2):115-39. doi: 10.1007/s10482-008-9284-5. Epub 2008 Oct 1. Antonie Van Leeuwenhoek. 2009. PMID: 18830684 Review.
-
The biological chemistry of the transition metal "transportome" of Cupriavidus metallidurans.Metallomics. 2016 May 1;8(5):481-507. doi: 10.1039/c5mt00320b. Metallomics. 2016. PMID: 27065183 Review.
Cited by
-
Antisense transcription is associated with expression of metal resistance determinants in Cupriavidus metallidurans CH34.Metallomics. 2024 Dec 2;16(12):mfae057. doi: 10.1093/mtomcs/mfae057. Metallomics. 2024. PMID: 39562278 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

