Control of spatio-temporal patterning via cell growth in a multicellular synthetic gene circuit
- PMID: 39562554
- PMCID: PMC11577002
- DOI: 10.1038/s41467-024-53078-8
Control of spatio-temporal patterning via cell growth in a multicellular synthetic gene circuit
Abstract
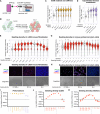
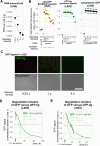
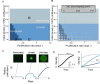
A major goal in synthetic development is to build gene regulatory circuits that control patterning. In natural development, an interplay between mechanical and chemical communication shapes the dynamics of multicellular gene regulatory circuits. For synthetic circuits, how non-genetic properties of the growth environment impact circuit behavior remains poorly explored. Here, we first describe an occurrence of mechano-chemical coupling in synthetic Notch (synNotch) patterning circuits: high cell density decreases synNotch-gated gene expression in different cellular systems in vitro. We then construct, both in vitro and in silico, a synNotch-based signal propagation circuit whose outcome can be regulated by cell density. Spatial and temporal patterning outcomes of this circuit can be predicted and controlled via modulation of cell proliferation, initial cell density, and/or spatial distribution of cell density. Our work demonstrates that synthetic patterning circuit outcome can be controlled via cellular growth, providing a means for programming multicellular circuit patterning outcomes.
© 2024. The Author(s).
Conflict of interest statement
Figures




References
-
- Thompson, D. W. On Growth and Form (Dover publication, New York, 1917).
-
- Fukaya, T. Dynamic regulation of anterior-posterior patterning genes in living Drosophila embryos. Curr. Biol.31, 2227–2236.e6 (2021). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- R35 GM138256/GM/NIGMS NIH HHS/United States
- CBET-2034495 RECODE/NSF | ENG/OAD | Division of Chemical, Bioengineering, Environmental, and Transport Systems (CBET)
- GM138256/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- CBET-2145528 CAREER/NSF | ENG/OAD | Division of Chemical, Bioengineering, Environmental, and Transport Systems (CBET)
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources

