ETV2 transcriptionally activates Rig1 gene expression and promotes reprogramming of the endothelial lineage
- PMID: 39562637
- PMCID: PMC11576751
- DOI: 10.1038/s41598-024-78115-w
ETV2 transcriptionally activates Rig1 gene expression and promotes reprogramming of the endothelial lineage
Abstract
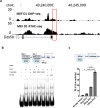
ETV2 is an essential transcription factor as Etv2 null murine embryos lack all vasculature, blood and are lethal early during embryogenesis. Previous studies have established that ETV2 functions as a pioneer factor and directly reprograms fibroblasts to endothelial cells. However, the underlying molecular mechanisms regulating this reprogramming process remain incompletely defined. In the present study, we examined the ETV2-RIG1 cascade as regulators that govern ETV2-mediated reprogramming. Mouse embryonic fibroblasts (MEFs) harboring an inducible ETV2 expression system were used to overexpress ETV2 and reprogram these somatic cells to the endothelial lineage. Single-cell RNA-seq from reprogrammed fibroblasts defined the induction of the transcriptional network involved in Rig1-like receptor signaling pathways. Studies using ChIP-seq, electrophoretic mobility shift assays, and transcriptional assays demonstrated that ETV2 was a direct upstream activator of Rig1 gene expression. We further demonstrated that the knockdown of Rig1 and separately, Nfκb1 using shRNA significantly reduced the efficiency of endothelial cell reprogramming. These results highlight that ETV2 reprograms fibroblasts to endothelial cells by directly activating RIG1. These findings extend our current understanding of the molecular mechanisms underlying ETV2-mediated reprogramming and will be important in the design of revascularization strategies for the treatment of ischemic tissues such as ischemic heart disease.
Keywords: ETV2; RIG1; Reprogramming.
© 2024. The Author(s).
Conflict of interest statement
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

