KMT5C leverages disorder to optimize cooperation with HP1 for heterochromatin retention
- PMID: 39562713
- PMCID: PMC11723951
- DOI: 10.1038/s44319-024-00320-5
KMT5C leverages disorder to optimize cooperation with HP1 for heterochromatin retention
Abstract
A defining feature of constitutive heterochromatin compartments is the heterochromatin protein-1 (HP1) family, whose members display fast internal mobility and rapid exchange with the surrounding nucleoplasm. Here, we describe a paradoxical state for the lysine methyltransferase KMT5C characterized by rapid internal diffusion but minimal nucleoplasmic exchange. This retentive behavior is conferred by sparse sequence features that constitute two modules tethered by an intrinsically disordered linker. While both modules harbor variant HP1 interaction motifs, the first comprises adjacent sequences that increase affinity using avidity. The second motif increases HP1 effective concentration to further enhance affinity in a context-dependent manner, which is evident using distinct heterochromatin recruitment strategies and heterologous linkers with defined conformational ensembles. Despite the linker sequence being highly divergent, it is under evolutionary constraint for functional length, suggesting conformational buffering can support cooperativity between modules across distant orthologs. Overall, we show that KMT5C has evolved a robust tethering strategy that uses minimal sequence determinants to harness highly dynamic HP1 proteins for retention within heterochromatin compartments.
Keywords: Heterochromatin Protein-1 (HP1); Intrinsic Disorder; Lysine Methyltransferase 5C (KMT5C); Protein Compartmentalization; Protein Dynamics.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures


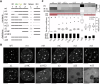


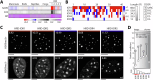




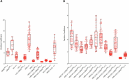
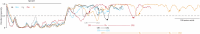
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 - PubMed
-
- Arganda-Carreras I, Kaynig V, Rueden C, Eliceiri KW, Schindelin J, Cardona A, Sebastian Seung H (2017) Trainable Weka segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33:2424–2426 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

