Polycomb group protein Mel18 inhibits hematopoietic stem cell self-renewal through repressing the transcription of self-renewal and proliferation genes
- PMID: 39562720
- PMCID: PMC12050008
- DOI: 10.1038/s41375-024-02462-w
Polycomb group protein Mel18 inhibits hematopoietic stem cell self-renewal through repressing the transcription of self-renewal and proliferation genes
Abstract
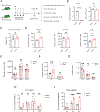
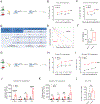
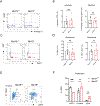
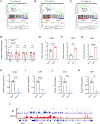
Polycomb group (PcG) proteins play important roles in hematopoietic stem cell (HSC) self-renewal. Mel18 and Bmi1 are homologs of the PCGF subunit within the Polycomb repressive complex 1 (PRC1). Bmi1 (PCGF4) enhances HSC self-renewal and promotes terminal differentiation. However, the role of Mel18 (PCGF2) in hematopoiesis is not fully understood and how Mel18 regulates gene transcription in HSCs remains elusive. We found that acute deletion of Mel18 in the hematopoietic compartment significantly increased the frequency of functional HSCs in the bone marrow. Furthermore, we demonstrate that Mel18 inhibits HSC self-renewal and proliferation. RNA-seq studies revealed that HSC self-renewal and proliferation gene signatures are enriched in Mel18-/- hematopoietic stem and progenitors (HSPCs) compared to Mel18+/+ HSPCs. Notably, ATAC-seq revealed increased chromatin accessibility at genes important for HSC self-renewal, whereas CUT&RUN showed decreased enrichment of H2AK119ub1 at genes important for proliferation, leading to increased expression of both Hoxb4 and Cdk4 in Mel18-/- HSPCs. Thus, we demonstrate that Mel18 inhibits hematopoietic stem cell self-renewal through repressing the transcription of genes important for HSC self-renewal and proliferation.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All methods were performed in accordance with the relevant guidelines and regulations. We have complied with all the relevant ethical regulations for animal testing and research. All animal-related experiments have received ethical approval from both Indiana and Northwestern University Institutional Animal Care and Use Committee (IACUC).
Figures

References
-
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, et al. Biology of Hematopoietic Stem Cells and Progenitors: Implications for Clinical Application. Annu Rev Immunol. 2003;21:759–806. - PubMed
-
- Attar EC, Scadden DT. Regulation of hematopoietic stem cell growth. Leukemia. 2004;18:1760–8. - PubMed
-
- Sorrentino BP. Clinical strategies for expansion of haematopoietic stem cells. Nat Rev Immunol. 2004;4(11):878–88. - PubMed
-
- Walasek MA, van Os R, de Haan G. Hematopoietic stem cell expansion: challenges and opportunities. Ann N Y Acad Sci. 2012;1266:138–50. - PubMed
-
- Akala OO, Clarke MF. Hematopoietic stem cell self-renewal. Curr Opin Genet Dev. 2006; 16:496–501. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

