Global atlas of predicted functional domains in Legionella pneumophila Dot/Icm translocated effectors
- PMID: 39562741
- PMCID: PMC11696984
- DOI: 10.1038/s44320-024-00076-z
Global atlas of predicted functional domains in Legionella pneumophila Dot/Icm translocated effectors
Abstract
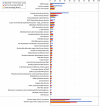
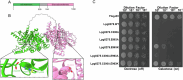
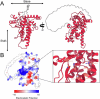
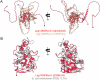
Legionella pneumophila utilizes the Dot/Icm type IVB secretion system to deliver hundreds of effector proteins inside eukaryotic cells to ensure intracellular replication. Our understanding of the molecular functions of the largest pathogenic arsenal known to the bacterial world remains incomplete. By leveraging advancements in 3D protein structure prediction, we provide a comprehensive structural analysis of 368 L. pneumophila effectors, representing a global atlas of predicted functional domains summarized in a database ( https://pathogens3d.org/legionella-pneumophila ). Our analysis identified 157 types of diverse functional domains in 287 effectors, including 159 effectors with no prior functional annotations. Furthermore, we identified 35 cryptic domains in 30 effector models that have no similarity with experimentally structurally characterized proteins, thus, hinting at novel functionalities. Using this analysis, we demonstrate the activity of thirteen functional domains, including three cryptic domains, predicted in L. pneumophila effectors to cause growth defects in the Saccharomyces cerevisiae model system. This illustrates an emerging strategy of exploring synergies between predictions and targeted experimental approaches in elucidating novel effector activities involved in infection.
Keywords: Legionella pneumophila; Bacterial Effectors; Cryptic Domains; Protein Modeling; Yeast Toxicity.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures





References
-
- Alexander JAN, Locher KP (2023) Emerging structural insights into C-type glycosyltransferases. Curr Opin Struct Biol 79:102547 - PubMed
MeSH terms
Substances
Grants and funding
- 75N93022C00035/AI/NIAID NIH HHS/United States
- MRI-2215705/National Science Foundation (NSF)
- MRI-1429826/National Science Foundation (NSF)
- HHSN272201700060C/AI/NIAID NIH HHS/United States
- S10 OD016290/OD/NIH HHS/United States
- PJT-162256/Canadian Government | CIHR | Canadian Institutes of Health Research - Antimicrobial Resistance Research Initiative
- RGPIN-2017-04878/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
- 1S10OD016290-01A1/HHS | NIH | NIAID | Division of Microbiology and Infectious Diseases (DMID)

