Drug-target interaction prediction by integrating heterogeneous information with mutual attention network
- PMID: 39563226
- PMCID: PMC11577831
- DOI: 10.1186/s12859-024-05976-3
Drug-target interaction prediction by integrating heterogeneous information with mutual attention network
Abstract
Background: Identification of drug-target interactions is an indispensable part of drug discovery. While conventional shallow machine learning and recent deep learning methods based on chemogenomic properties of drugs and target proteins have pushed this prediction performance improvement to a new level, these methods are still difficult to adapt to novel structures. Alternatively, large-scale biological and pharmacological data provide new ways to accelerate drug-target interaction prediction.
Methods: Here, we propose DrugMAN, a deep learning model for predicting drug-target interaction by integrating multiplex heterogeneous functional networks with a mutual attention network (MAN). DrugMAN uses a graph attention network-based integration algorithm to learn network-specific low-dimensional features for drugs and target proteins by integrating four drug networks and seven gene/protein networks collected by a certain screening conditions, respectively. DrugMAN then captures interaction information between drug and target representations by a mutual attention network to improve drug-target prediction.
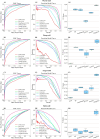
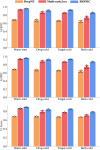
Results: DrugMAN achieved the best performance compared with cheminformation-based methods SVM, RF, DeepPurpose and network-based deep learing methods DTINet and NeoDT in four different scenarios, especially in real-world scenarios. Compared with SVM, RF, deepurpose, DTINet, and NeoDT, DrugMAN showed the smallest decrease in AUROC, AUPRC, and F1-Score from warm-start to Both-cold scenarios. This result is attributed to DrugMAN's learning from heterogeneous data and indicates that DrugMAN has a good generalization ability. Taking together, DrugMAN spotlights heterogeneous information to mine drug-target interactions and can be a powerful tool for drug discovery and drug repurposing.
Keywords: Deep learning; Drug discovery; Drug–target interaction; Heterogeneous network; Self-attention.
© 2024. The Author(s).
Conflict of interest statement
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

