Private detection of relatives in forensic genomics using homomorphic encryption
- PMID: 39563334
- PMCID: PMC11575431
- DOI: 10.1186/s12920-024-02037-9
Private detection of relatives in forensic genomics using homomorphic encryption
Abstract
Background: Forensic analysis heavily relies on DNA analysis techniques, notably autosomal Single Nucleotide Polymorphisms (SNPs), to expedite the identification of unknown suspects through genomic database searches. However, the uniqueness of an individual's genome sequence designates it as Personal Identifiable Information (PII), subjecting it to stringent privacy regulations that can impede data access and analysis, as well as restrict the parties allowed to handle the data. Homomorphic Encryption (HE) emerges as a promising solution, enabling the execution of complex functions on encrypted data without the need for decryption. HE not only permits the processing of PII as soon as it is collected and encrypted, such as at a crime scene, but also expands the potential for data processing by multiple entities and artificial intelligence services.
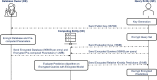
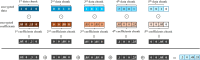
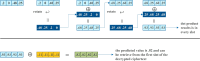
Methods: This study introduces HE-based privacy-preserving methods for SNP DNA analysis, offering a means to compute kinship scores for a set of genome queries while meticulously preserving data privacy. We present three distinct approaches, including one unsupervised and two supervised methods, all of which demonstrated exceptional performance in the iDASH 2023 Track 1 competition.
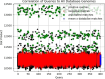
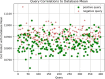
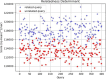
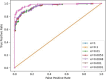
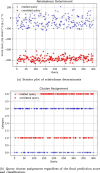
Results: Our HE-based methods can rapidly predict 400 kinship scores from an encrypted database containing 2000 entries within seconds, capitalizing on advanced technologies like Intel AVX vector extensions, Intel HEXL, and Microsoft SEAL HE libraries. Crucially, all three methods achieve remarkable accuracy levels (ranging from 96% to 100%), as evaluated by the auROC score metric, while maintaining robust 128-bit security. These findings underscore the transformative potential of HE in both safeguarding genomic data privacy and streamlining precise DNA analysis.
Conclusions: Results demonstrate that HE-based solutions can be computationally practical to protect genomic privacy during screening of candidate matches for further genealogy analysis in Forensic Genetic Genealogy (FGG).
Keywords: Data privacy; Genomic database; Homomorphic encryption; Secure query.
© 2024. The Author(s).
Conflict of interest statement
Figures












References
-
- GEDmatchPRO. GEDmatch PRO. 2023. https://pro.gedmatch.com/user/login?destination. Accessed 29 Dec 2023
-
- FamilyTreeDNA. DNA Testing for Ancestry and Genealogy | Family Tree DNA. 2023. https://www.familytreedna.com/. Accessed 29 Dec 2023
-
- DNASolves. DNASolves. 2023. https://dnasolves.com/. Accessed 29 Dec 2023.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

