Distinct Inflammatory Programs Underlie the Intramuscular Lipid Nanoparticle Response
- PMID: 39563529
- PMCID: PMC12180297
- DOI: 10.1021/acsnano.4c08490
Distinct Inflammatory Programs Underlie the Intramuscular Lipid Nanoparticle Response
Abstract
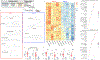
Developments in mRNA/lipid nanoparticle (LNP) technology have advanced the fields of vaccinology and gene therapy, raising questions about immunogenicity. While some mRNA/LNPs generate an adjuvant-like environment in muscle tissue, other mRNA/LNPs are distinct in their capacity for multiple rounds of therapeutic delivery. We evaluate the adjuvancy of components of mRNA/LNPs by phenotyping cellular infiltrate at injection sites, tracking uptake by immune cells, and assessing the inflammatory state. Delivery of 9 common, but chemically distinct, LNPs to muscle revealed two classes of inflammatory gene expression programs: inflammatory (Class A) and noninflammatory (Class B). We find that intramuscular injection with Class A, but not Class B, empty LNPs (eLNPs) induce robust neutrophil infiltration into muscle within 2 h and a diverse myeloid population within 24 h. Single-cell RNA sequencing revealed SM-102-mediated expression of inflammatory chemokines by myeloid infiltrates within muscle 1 day after injection. Surprisingly, we found direct transfection of muscle infiltrating myeloid cells and splenocytes 24 h after intramuscular mRNA/LNP administration. Transfected myeloid cells within the muscle exhibit an activated phenotype 24 h after injection. Similarly, directly transfected splenic lymphocytes and dendritic cells (DCs) are differentially activated by Class A or Class B containing mRNA/LNP. Within the splenic DC compartment, type II conventional DCs (cDC2s) are directly transfected and activated by Class A mRNA/LNP. Together, we show that mRNA and LNPs work synergistically to provide the necessary innate immune stimuli required for effective vaccination. Importantly, this work provides a design framework for vaccines and therapeutics alike.
Keywords: cancer vaccine; gene therapy; innate immunity; ionizable lipids; lipid nanoparticles; mRNA therapeutics; mRNA vaccines.
Conflict of interest statement
Notes
The authors declare no competing financial interest.
Figures
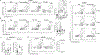


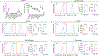
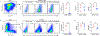
References
-
- Baden LR; El Sahly HM; Essink B; Kotloff K; Frey S; Novak R; Diemert D; Spector SA; Rouphael N; Creech CB; McGettigan J; Khetan S; Segall N; Solis J; Brosz A; Fierro C; Schwartz H; Neuzil K; Corey L; Gilbert P; Janes H; Follmann D; Marovich M; Mascola J; Polakowski L; Ledgerwood J; Graham BS; Bennett H; Pajon R; Knightly C; Leav B; Deng W; Zhou H; Han S; Ivarsson M; Miller J; Zaks T. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384 (5), 403–416. - PMC - PubMed
-
- Polack FP; Thomas SJ; Kitchin N; Absalon J; Gurtman A; Lockhart S; Perez JL; Marc GP; Moreira ED; Zerbini C; Bailey R; Swanson KA; Roychoudhury S; Koury K; Li P; Kalina WV; Cooper D; Frenck RW; Hammitt LL; Türeci Ö; Nell H; Schaefer A; Ünal S; Tresnan DB; Mather S; Dormitzer PR; Şahin U; Jansen KU; Gruber WC. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383 (27), 2603–2615. - PMC - PubMed
-
- Kim W; Zhou JQ; Horvath SC; Schmitz AJ; Sturtz AJ; Lei T; Liu Z; Kalaidina E; Thapa M; Alsoussi WB; Haile A; Klebert MK; Suessen T; Parra-Rodriguez L; Mudd PA; Whelan SPJ; Middleton WD; Teefey SA; Pusic I; O’Halloran JA; Presti RM; Turner JS; Ellebedy AH. Germinal Centre-Driven Maturation of B Cell Response to mRNA Vaccination. Nature 2022, 604 (7904), 141–145. - PMC - PubMed
-
- Turner JS; O’Halloran JA; Kalaidina E; Kim W; Schmitz AJ; Zhou JQ; Lei T; Thapa M; Chen RE; Case JB; Amanat F; Rauseo AM; Haile A; Xie X; Klebert MK; Suessen T; Middleton WD; Shi P-Y; Krammer F; Teefey SA; Diamond MS; Presti RM; Ellebedy AH. SARS-CoV-2 mRNA Vaccines Induce Persistent Human Germinal Centre Responses. Nature 2021, 596 (7870), 109–113. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

