Exploring and Validating the Mechanism of Ulinastatin in the Treatment of Sepsis-Associated Encephalopathy Based on Transcriptome Sequencing
- PMID: 39564549
- PMCID: PMC11573691
- DOI: 10.2147/JIR.S488400
Exploring and Validating the Mechanism of Ulinastatin in the Treatment of Sepsis-Associated Encephalopathy Based on Transcriptome Sequencing
Abstract
Purpose: Sepsis can induce sepsis-associated encephalopathy (SAE), with Ulinastatin (UTI) serving a critical anti-inflammatory role. This study aimed to identify the hub genes in an SAE mouse model following UTI intervention and investigate the underlying molecular mechanisms.
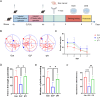
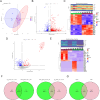
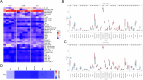
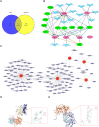
Materials and methods: Through differential expression analysis to obtain differentially expressed genes (DEGs), ie, UTI vs CLP (DEGs1) and Con vs CLP (DEGs2). After taking the intersection of the genes with opposite differential trends in these two parts and immune-related genes (IRGs), DE-IRGs were obtained. Hub genes in the protein-protein interaction (PPI) network were then determined using six algorithms from the Cytohubba plugin in Cytoscape. Gene set enrichment analysis (GSEA) was employed to explore the functional relevance of these hub genes. Additionally, the immune microenvironment across the three groups was compared, and hub gene-related drugs were predicted using an online database. Finally, qRT-PCR was used to validate the expression of the hub genes in hippocampal tissue from CLP mice.
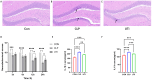
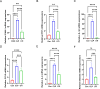
Results: RNA sequencing obtained 864 differentially expressed genes (DEGs) (CLP vs Con) and 279 DEGs (UTI vs CLP). Taking the intersection of DEGs with opposite expression trends yielded 165 DEGs. Six key genes (ICAM - 1, IRF7, IL - 1β, CCL2, IL - 6 and SOCS3) were screened by six algorithms. Immune infiltration analysis found that Treg cells were reversed after treatment with UTI in the diseased state. A total of 106 hub - gene - related drugs were predicted, among which BINDARIT - CCL2 and LIFITEGRAST - ICAM1 showed particularly high affinities. The qRT - PCR verification results were consistent with the sequencing results.
Conclusion: In conclusion, ICAM-1, IRF7, IL-1β, CCL2, IL-6, and SOCS3 were identified as potential therapeutic targets in SAE mice treated with UTI. This study offers theoretical support for UTI as a treatment option for SAE.
Keywords: RNA sequencing; Treg cells; immune; sepsis-associated encephalopathy; ulinastatin.
© 2024 Hu et al.
Conflict of interest statement
Wen Hu, Xiaoyuan Zhang, Zhen Wu, and Yushan Luo are co-first authors for this study. The authors declare that they have no competing interests in this work.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

