Whole genome sequencing revealed high proportions of ST152 MRSA among clinical Staphylococcus aureus isolates from ten hospitals in Ghana
- PMID: 39565128
- PMCID: PMC11656792
- DOI: 10.1128/msphere.00446-24
Whole genome sequencing revealed high proportions of ST152 MRSA among clinical Staphylococcus aureus isolates from ten hospitals in Ghana
Abstract
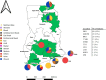
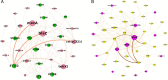
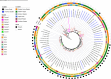
Previous studies in Ghana indicated low prevalence of methicillin-resistant Staphylococcus aureus (MRSA) and predominance of ST152 methicillin-susceptible S. aureus (MSSA) among clinical isolates. ST152 MRSA clones are associated with severe infections and epidemics. Using whole genome sequencing (WGS), 159 S. aureus isolated from clinical sources (wound, blood, urine, ear, abscess, umbilical cord, eye, vaginal samples, and others) from 10 hospitals across Ghana were investigated. mecA (gene for methicillin resistance) was detected in 38% of the isolates. Panton-Valentine leucocidin toxin (PVL) gene occurred in 65% isolates, with 84% of the MRSA's harboring the PVL gene. ST152 was the major clone, with 74% harboring the mecA gene. Other MRSA clones detected were ST5, ST5204, ST852, and ST1. MSSA clones included ST3249, ST152, ST5, ST1, and ST8. Twenty-three genes encoding resistance to 12 antimicrobial classes were observed with blaZ (97%) being the most prevalent. Other predominant resistance genes included tetK (46%), cat (42%), and dfrG (36%) encoding resistance for tetracyclines, phenicols, and diaminopyrimidine, respectively. Virulence genes for enterotoxins, biofilms, toxic-shock-syndrome toxins, hemolysins, and leukotoxins were also detected. Phylogenetic analysis revealed a shift in the dominant clone from MSSA ST152 to MRSA ST152 over the past decade. The study provides valuable insights into the genomic content of S. aureus from clinical sources in Ghana. The finding of ST152 MRSA in high numbers suggests a shifting epidemiological landscape of these pathogens and continuous surveillance using robust tools like WGS is needed to monitor the rise and spread of these epidemic clones in the country.IMPORTANCESince its emergence in 1959, MRSA has been a significant public health concern, causing infections in both clinical and community settings. Patients with MRSA-related infections experience higher mortality rates due to its ability to evade antimicrobials and immune defenses. In Ghana, understanding the molecular epidemiology of MRSA has been hindered by the lack of appropriate laboratory infrastructure and the limited capacity for molecular data analysis. This study, the largest genomic study of S. aureus in Ghana, addresses this gap by utilizing whole genome sequencing to examine the diversity of circulating S. aureus strains from 10 hospitals. Our findings highlight the predominance of pandemic clones, particularly ST152, and the notable transition of ST152 MSSA to ST152 MRSA over the past decade. The findings from this study supports AMR surveillance efforts in Ghana and emphasize the importance of implementing genomic surveillance using WGS to comprehensively monitor the rise and spread of multi-drug-resitant organisms such as MRSA in the country.
Keywords: Africa; ST152 methicillin-resistant S. aureus; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Molecular characteristics of community-acquired methicillin-resistant Staphylococcus aureus in Hokkaido, northern main island of Japan: identification of sequence types 6 and 59 Panton-Valentine leucocidin-positive community-acquired methicillin-resistant Staphylococcus aureus.Microb Drug Resist. 2011 Jun;17(2):241-50. doi: 10.1089/mdr.2010.0136. Epub 2011 Mar 13. Microb Drug Resist. 2011. PMID: 21395449
-
Clinical and Molecular Epidemiology of an Emerging Panton-Valentine Leukocidin-Positive ST5 Methicillin-Resistant Staphylococcus aureus Clone in Northern Australia.mSphere. 2021 Feb 10;6(1):e00651-20. doi: 10.1128/mSphere.00651-20. mSphere. 2021. PMID: 33568451 Free PMC article.
-
Local circulating clones of Staphylococcus aureus in Ecuador.Braz J Infect Dis. 2016 Nov-Dec;20(6):525-533. doi: 10.1016/j.bjid.2016.08.006. Epub 2016 Sep 13. Braz J Infect Dis. 2016. PMID: 27638417 Free PMC article.
-
Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus.Infect Genet Evol. 2014 Jan;21:563-74. doi: 10.1016/j.meegid.2013.04.030. Epub 2013 May 3. Infect Genet Evol. 2014. PMID: 23648426 Free PMC article. Review.
-
Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power.Curr Opin Microbiol. 2007 Oct;10(5):428-35. doi: 10.1016/j.mib.2007.08.003. Epub 2007 Oct 24. Curr Opin Microbiol. 2007. PMID: 17921044 Free PMC article. Review.
Cited by
-
Case Report: Fatal Necrotizing Pneumonia by Exfoliative Toxin etE2-Producing Staphylococcus aureus Belonging to MLST ST152 in The Netherlands.Microorganisms. 2025 Jul 9;13(7):1618. doi: 10.3390/microorganisms13071618. Microorganisms. 2025. PMID: 40732126 Free PMC article.
-
Methicillin-resistant Staphylococcus aureus in Saudi Arabia: genomic evidence of recent clonal expansion and plasmid-driven resistance dissemination.Front Microbiol. 2025 Jun 13;16:1602985. doi: 10.3389/fmicb.2025.1602985. eCollection 2025. Front Microbiol. 2025. PMID: 40584034 Free PMC article.
References
-
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi:10.1016/S1473-3099(17)30753-3 - DOI - PubMed
-
- Aung MS, Urushibara N, Kawaguchiya M, Sumi A, Shinagawa M, Takahashi S, Kobayashi N. 2019. Clonal diversity and genetic characteristics of methicillin-resistant Staphylococcus aureus isolates from a tertiary care hospital in Japan. Microb Drug Resist 25:1164–1175. doi:10.1089/mdr.2018.0468 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

