ECOD: integrating classifications of protein domains from experimental and predicted structures
- PMID: 39565196
- PMCID: PMC11701565
- DOI: 10.1093/nar/gkae1029
ECOD: integrating classifications of protein domains from experimental and predicted structures
Abstract
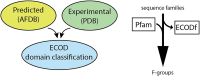
The evolutionary classification of protein domains (ECOD) classifies protein domains using a combination of sequence and structural data (http://prodata.swmed.edu/ecod). Here we present the culmination of our previous efforts at classifying domains from predicted structures, principally from the AlphaFold Database (AFDB), by integrating these domains with our existing classification of PDB structures. This combined classification includes both domains from our previous, purely experimental, classification of domains as well as domains from our provisional classification of 48 proteomes in AFDB predicted from model organisms and organisms of concern to global health. ECOD classifies over 1.8 M domains from over 1000 000 proteins collectively deposited in the PDB and AFDB. Additionally, we have changed the F-group classification reference used for ECOD, deprecating our original ECODf library and instead relying on direct collaboration with the Pfam sequence family database to inform our classification. Pfam provides similar coverage of ECOD with family classification while being more accurate and less redundant. By eliminating duplication of effort, we can improve both classifications. Finally, we discuss the initial deployment of DrugDomain, a database of domain-ligand interactions, on ECOD and discuss future plans.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
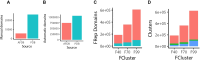




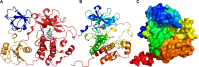
Similar articles
-
ECOD: new developments in the evolutionary classification of domains.Nucleic Acids Res. 2017 Jan 4;45(D1):D296-D302. doi: 10.1093/nar/gkw1137. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899594 Free PMC article.
-
A sequence family database built on ECOD structural domains.Bioinformatics. 2018 Sep 1;34(17):2997-3003. doi: 10.1093/bioinformatics/bty214. Bioinformatics. 2018. PMID: 29659718 Free PMC article.
-
ECOD: an evolutionary classification of protein domains.PLoS Comput Biol. 2014 Dec 4;10(12):e1003926. doi: 10.1371/journal.pcbi.1003926. eCollection 2014 Dec. PLoS Comput Biol. 2014. PMID: 25474468 Free PMC article.
-
Classification of proteins with shared motifs and internal repeats in the ECOD database.Protein Sci. 2016 Jul;25(7):1188-203. doi: 10.1002/pro.2893. Epub 2016 Feb 21. Protein Sci. 2016. PMID: 26833690 Free PMC article. Review.
-
Pfam 10 years on: 10,000 families and still growing.Brief Bioinform. 2008 May;9(3):210-9. doi: 10.1093/bib/bbn010. Epub 2008 Mar 15. Brief Bioinform. 2008. PMID: 18344544 Review.
Cited by
-
The 2025 Nucleic Acids Research database issue and the online molecular biology database collection.Nucleic Acids Res. 2025 Jan 6;53(D1):D1-D9. doi: 10.1093/nar/gkae1220. Nucleic Acids Res. 2025. PMID: 39658041 Free PMC article.
-
Gardnerella fibrinogen-binding protein as a candidate adherence factor.Front Cell Infect Microbiol. 2025 May 8;15:1556232. doi: 10.3389/fcimb.2025.1556232. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40406528 Free PMC article.
-
Assessing Structural Classification Using AlphaFold2 Models Through ECOD-Based Comparative Analysis.Proteins. 2025 Sep;93(9):1571-1585. doi: 10.1002/prot.26828. Epub 2025 Apr 19. Proteins. 2025. PMID: 40251890 Free PMC article.
-
Leveraging AI to explore structural contexts of post-translational modifications in drug binding.J Cheminform. 2025 May 4;17(1):67. doi: 10.1186/s13321-025-01019-y. J Cheminform. 2025. PMID: 40320551 Free PMC article.
-
DrugDomain 2.0: comprehensive database of protein domains-ligands/drugs interactions across the whole Protein Data Bank.bioRxiv [Preprint]. 2025 Jul 7:2025.07.03.663025. doi: 10.1101/2025.07.03.663025. bioRxiv. 2025. PMID: 40672152 Free PMC article. Preprint.
References
-
- Murzin A.G., Brenner S.E., Hubbard T., Chothia C.. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995; 247:536–540. - PubMed

