[SNG2], a prion form of Cut4/Apc1, confers non-Mendelian inheritance of heterochromatin silencing defect in fission yeast
- PMID: 39565210
- PMCID: PMC11662930
- DOI: 10.1093/nar/gkae1136
[SNG2], a prion form of Cut4/Apc1, confers non-Mendelian inheritance of heterochromatin silencing defect in fission yeast
Abstract
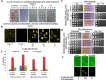
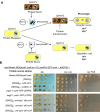
Prions represent epigenetic regulator proteins that can self-propagate their structure and confer their misfolded structure and function on normally folded proteins. Like the mammalian prion PrPSc, prions also occur in fungi. While a few prions, like Swi1, affect gene expression, none are shown to affect heterochromatin structure and function. In fission yeast and metazoans, histone methyltransferase Clr4/Suv39 causes H3-Lys9 methylation, which is bound by the chromodomain protein Swi6/HP1 to assemble heterochromatin. Earlier, we showed that sng2-1 mutation in the Cut4 subunit of anaphase-promoting complex abrogates heterochromatin structure due to defective binding and recruitment of Swi6. Here, we demonstrate that the Cut4p forms a non-canonical prion form, designated as [SNG2], which abrogates heterochromatin silencing. [SNG2] exhibits various prion-like properties, e.g. non-Mendelian inheritance, requirement of Hsp proteins for its propagation, de novo generation upon cut4 overexpression, reversible curing by guanidine, cytoplasmic inheritance and formation of infectious protein aggregates, which are dissolved upon overexpression of hsp genes. Interestingly, [SNG2] prion imparts an enhanced tolerance to stress conditions, supporting its role in promoting cell survival under environmental stress during evolution.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures







References
-
- Prusiner S.B. Novel proteinaceous infectious particles cause scrapie. Science. 1982; 216:136–144. - PubMed
-
- Oesch B., Westaway D., Wälchli M., McKinley M.P., Kent S.B., Aebersold R., Barry R.A., Tempst P., Teplow D.B., Hood L.E.. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 1985; 40:735–746. - PubMed
-
- Moore R.C., Hope J., McBride P.A., McConnell I., Selfridge J., Melton D.W., Manson J.C.. Mice with gene targeted prion protein alterations show that Prnp, Sinc and Prni are congruent. Nat. Genet. 1998; 18:118–125. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

