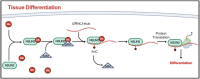
Glucose binds and activates NSUN2 to promote translation and epidermal differentiation
- PMID: 39565212
- PMCID: PMC11662651
- DOI: 10.1093/nar/gkae1097
Glucose binds and activates NSUN2 to promote translation and epidermal differentiation
Abstract
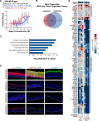
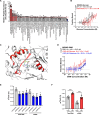
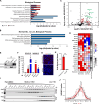
Elevations in intracellular glucose concentrations are essential for epithelial cell differentiation by mechanisms that are not fully understood. Glucose has recently been found to directly bind several proteins to alter their functions to enhance differentiation. Among the newly identified glucose-binding proteins is NSUN2, an RNA-binding protein that we identified as indispensable for epidermal differentiation. Glucose was found to bind conserved sequences within NSUN2, enhancing its binding to S-adenosyl-L-methionine and boosting its enzymatic activity. Additionally, glucose enhanced NSUN2's proximity to proteins involved in mRNA translation, with NSUN2 modulating global messenger RNA (mRNA) translation, particularly that of key pro-differentiation mRNAs containing m5C modifications, such as GRHL3. Glucose thus engages diverse molecular mechanisms beyond its energetic roles to facilitate cellular differentiation processes.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

