Spatial and temporal distribution of ribosomes in single cells reveals aging differences between old and new daughters of Escherichia coli
- PMID: 39565213
- PMCID: PMC11578582
- DOI: 10.7554/eLife.89543
Spatial and temporal distribution of ribosomes in single cells reveals aging differences between old and new daughters of Escherichia coli
Abstract
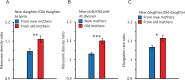
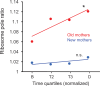
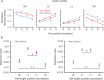
Lineages of rod-shaped bacteria such as Escherichia coli exhibit a temporal decline in elongation rate in a manner comparable to cellular or biological aging. The effect results from the production of asymmetrical daughters, one with a lower elongation rate, by the division of a mother cell. The slower daughter compared to the faster daughter, denoted respectively as the old and new daughters, has more aggregates of damaged proteins and fewer expressed gene products. We have examined further the degree of asymmetry by measuring the density of ribosomes between old and new daughters and between their poles. We found that ribosomes were denser in the new daughter and also in the new pole of the daughters. These ribosome patterns match the ones we previously found for expressed gene products. This outcome suggests that the asymmetry is not likely to result from properties unique to the gene expressed in our previous study, but rather from a more fundamental upstream process affecting the distribution of ribosomal abundance. Because damage aggregates and ribosomes are both more abundant at the poles of E. coli cells, we suggest that competition for space between the two could explain the reduced ribosomal density in old daughters. Using published values for aggregate sizes and the relationship between ribosomal number and elongation rates, we show that the aggregate volumes could in principle displace quantitatively the amount of ribosomes needed to reduce the elongation rate of the old daughters.
Keywords: E. coli; aging; asymmetry; damage; evolutionary biology; ribosomes; single cell.
© 2023, Chao, Chan et al.
Conflict of interest statement
LC, CC, CS, UR No competing interests declared
Figures




Update of
- doi: 10.1101/2023.06.21.545935
- doi: 10.7554/eLife.89543.1
- doi: 10.7554/eLife.89543.2
Similar articles
-
Allocation of gene products to daughter cells is determined by the age of the mother in single Escherichia coli cells.Proc Biol Sci. 2020 May 13;287(1926):20200569. doi: 10.1098/rspb.2020.0569. Epub 2020 May 6. Proc Biol Sci. 2020. PMID: 32370668 Free PMC article.
-
Spatial Distribution and Ribosome-Binding Dynamics of EF-P in Live Escherichia coli.mBio. 2017 Jun 6;8(3):e00300-17. doi: 10.1128/mBio.00300-17. mBio. 2017. PMID: 28588135 Free PMC article.
-
Progressive decline in old pole gene expression signal enhances phenotypic heterogeneity in bacteria.Sci Adv. 2024 Nov 8;10(45):eadp8784. doi: 10.1126/sciadv.adp8784. Epub 2024 Nov 8. Sci Adv. 2024. PMID: 39514668 Free PMC article.
-
Is Escherichia coli getting old?Bioessays. 2005 Aug;27(8):770-4. doi: 10.1002/bies.20271. Bioessays. 2005. PMID: 16015607 Review.
-
[tRNA-binding centers of Escherichia coli ribosomes and their structural organization].Mol Biol (Mosk). 1984 Sep-Oct;18(5):1194-207. Mol Biol (Mosk). 1984. PMID: 6209546 Review. Russian.
Cited by
-
Nonequilibrium polysome dynamics promote chromosome segregation and its coupling to cell growth in Escherichia coli.Elife. 2025 Jun 24;14:RP104276. doi: 10.7554/eLife.104276. Elife. 2025. PMID: 40552714 Free PMC article.
-
A link between aging and persistence.Antimicrob Agents Chemother. 2025 Apr 2;69(4):e0131324. doi: 10.1128/aac.01313-24. Epub 2025 Feb 21. Antimicrob Agents Chemother. 2025. PMID: 39982072 Free PMC article.
-
Single-cell heterogeneity in ribosome content and the consequences for the growth laws.bioRxiv [Preprint]. 2024 Oct 8:2024.04.19.590370. doi: 10.1101/2024.04.19.590370. bioRxiv. 2024. PMID: 38895328 Free PMC article. Preprint.
-
Nonequilibrium polysome dynamics promote chromosome segregation and its coupling to cell growth in Escherichia coli.bioRxiv [Preprint]. 2025 Mar 15:2024.10.08.617237. doi: 10.1101/2024.10.08.617237. bioRxiv. 2025. Update in: Elife. 2025 Jun 24;14:RP104276. doi: 10.7554/eLife.104276. PMID: 40161845 Free PMC article. Updated. Preprint.
References
-
- Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo J-M, Hardt W-D, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan M-W, Tenson T, Van Melderen L, Zinkernagel A. Definitions and guidelines for research on antibiotic persistence. Nature Reviews. Microbiology. 2019;17:441–448. doi: 10.1038/s41579-019-0196-3. - DOI - PMC - PubMed
-
- Cabrera JE, Cagliero C, Quan S, Squires CL, Jin DJ. Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: examining the effect of transcription on nucleoid structure in the absence of transertion. Journal of Bacteriology. 2009;191:4180–4185. doi: 10.1128/JB.01707-08. - DOI - PMC - PubMed

