Zika virus remodels and hijacks IGF2BP2 ribonucleoprotein complex to promote viral replication organelle biogenesis
- PMID: 39565347
- PMCID: PMC11578589
- DOI: 10.7554/eLife.94347
Zika virus remodels and hijacks IGF2BP2 ribonucleoprotein complex to promote viral replication organelle biogenesis
Abstract
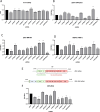
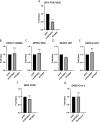
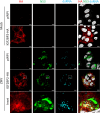
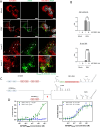
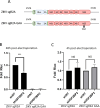
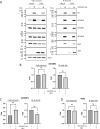
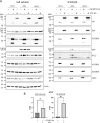
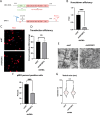
Zika virus (ZIKV) infection causes significant human disease that, with no approved treatment or vaccine, constitutes a major public health concern. Its life cycle entirely relies on the cytoplasmic fate of the viral RNA genome (vRNA) through a fine-tuned equilibrium between vRNA translation, replication, and packaging into new virions, all within virus-induced replication organelles (vROs). In this study, with an RNA interference (RNAi) mini-screening and subsequent functional characterization, we have identified insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) as a new host dependency factor that regulates vRNA synthesis. In infected cells, IGF2BP2 associates with viral NS5 polymerase and redistributes to the perinuclear viral replication compartment. Combined fluorescence in situ hybridization-based confocal imaging, in vitro binding assays, and immunoprecipitation coupled to RT-qPCR showed that IGF2BP2 directly interacts with ZIKV vRNA 3' nontranslated region. Using ZIKV sub-genomic replicons and a replication-independent vRO induction system, we demonstrated that IGF2BP2 knockdown impairs de novo vRO biogenesis and, consistently, vRNA synthesis. Finally, the analysis of immunopurified IGF2BP2 complex using quantitative mass spectrometry and RT-qPCR revealed that ZIKV infection alters the protein and RNA interactomes of IGF2BP2. Altogether, our data support that ZIKV hijacks and remodels the IGF2BP2 ribonucleoprotein complex to regulate vRO biogenesis and vRNA neosynthesis.
Keywords: IGF2BP2; RNA-binding proteins; Zika virus; cell biology; igf2bp2; infectious disease; microbiology; viral replication organelle; viral rna; zika virus.
© 2024, Mazeaud et al.
Conflict of interest statement
CM, SP, JO, HP, FC, ZR, AA, CB, AS, TP, CN, PS, LC No competing interests declared
Figures












Update of
- doi: 10.1101/2023.12.08.570783
- doi: 10.7554/eLife.94347.1
- doi: 10.7554/eLife.94347.2
References
-
- Anton A, Mazeaud C, Freppel W, Gilbert C, Tremblay N, Sow AA, Roy M, Rodrigue-Gervais IG, Chatel-Chaix L. Valosin-containing protein ATPase activity regulates the morphogenesis of Zika virus replication organelles and virus-induced cell death. Cellular Microbiology. 2021;23:e13302. doi: 10.1111/cmi.13302. - DOI - PubMed
-
- Bateman A, Martin M-J, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, Bye-A-Jee H, Coetzee R, Cukura A, Da Silva A, Denny P, Dogan T, Ebenezer T, Fan J, Castro LG, Garmiri P, Georghiou G, Gonzales L, Hatton-Ellis E, Hussein A, Ignatchenko A, Insana G, Ishtiaq R, Jokinen P, Joshi V, Jyothi D, Lock A, Lopez R, Luciani A, Luo J, Lussi Y, MacDougall A, Madeira F, Mahmoudy M, Menchi M, Mishra A, Moulang K, Nightingale A, Oliveira CS, Pundir S, Qi G, Raj S, Rice D, Lopez MR, Saidi R, Sampson J, Sawford T, Speretta E, Turner E, Tyagi N, Vasudev P, Volynkin V, Warner K, Watkins X, Zaru R, Zellner H, Bridge A, Poux S, Redaschi N, Aimo L, Argoud-Puy G, Auchincloss A, Axelsen K, Bansal P, Baratin D, Blatter M-C, Bolleman J, Boutet E, Breuza L, Casals-Casas C, de Castro E, Echioukh KC, Coudert E, Cuche B, Doche M, Dornevil D, Estreicher A, Famiglietti ML, Feuermann M, Gasteiger E, Gehant S, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hyka-Nouspikel N, Jungo F, Keller G, Kerhornou A, Lara V, Le Mercier P, Lieberherr D, Lombardot T, Martin X, Masson P, Morgat A, Neto TB, Paesano S, Pedruzzi I, Pilbout S, Pourcel L, Pozzato M, Pruess M, Rivoire C, Sigrist C, Sonesson K, Stutz A, Sundaram S, Tognolli M, Verbregue L, Wu CH, Arighi CN, Arminski L, Chen C, Chen Y, Garavelli JS, Huang H, Laiho K, McGarvey P, Natale DA, Ross K, Vinayaka CR, Wang Q, Wang Y, Yeh L-S, Zhang J, Ruch P, Teodoro D, The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. - DOI - PMC - PubMed
-
- Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cellular and Molecular Life Sciences. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- RGPIN-2022-03391/Natural Sciences and Engineering Research Council of Canada
- RGPIN-2016-05584/Natural Sciences and Engineering Research Council of Canada
- CFI-37155/Canada Foundation for Innovation
- CFI-41008/Canada Foundation for Innovation
- 2018-NC-205593/Fonds de recherche du Québec - Nature et technologies
- VirMScan/Bundesministerium für Bildung und Forschung
- SC 314/1-2/Deutsche Forschungsgemeinschaft
- PhD fellowship/Fonds de recherche du Québec - Nature et technologies
- PhD fellowship/Armand-Frappier Foundation
- PhD fellowship/Center of Excellence in Research on Orphan Diseases - Fondation Courtois
- CREATE postdoctoral fellowship award/Natural Sciences and Engineering Research Council of Canada
LinkOut - more resources
Full Text Sources
Medical
Research Materials

