Spc2 modulates substrate- and cleavage site-selection in the yeast signal peptidase complex
- PMID: 39565596
- PMCID: PMC11579918
- DOI: 10.1083/jcb.202211035
Spc2 modulates substrate- and cleavage site-selection in the yeast signal peptidase complex
Abstract
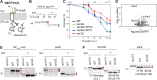
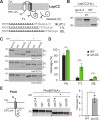
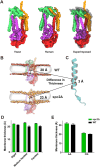
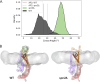
Secretory proteins are critically dependent on the correct processing of their signal sequence by the signal peptidase complex (SPC). This step, which is essential for the proper folding and localization of proteins in eukaryotic cells, is still not fully understood. In eukaryotes, the SPC comprises four evolutionarily conserved membrane subunits (Spc1-3 and Sec11). Here, we investigated the role of Spc2, examining SPC cleavage efficiency on various models and natural signal sequences in yeast cells depleted of or with mutations in Spc2. Our data show that discrimination between substrates and identification of the cleavage site by SPC is compromised when Spc2 is absent or mutated. Molecular dynamics simulation of the yeast SPC AlphaFold2-Multimer model indicates that membrane thinning at the center of SPC is reduced without Spc2, suggesting a molecular explanation for the altered substrate recognition properties of SPC lacking Spc2. These results provide new insights into the molecular mechanisms by which SPC governs protein biogenesis.
© 2024 Chung et al.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. P.C.T. Souza reported “Computer hours awarded to the GENCI project number 2022-A0120713456 and 2023-A0140713456 on the Jean-Zay (IDRIS), Adastra (CINES), and Joliot-Curie (TGCC) clusters of the French National Supercomputing Center (GENCI) and the computing time available at the IN2P3 computer cluster in Lyon are gratefully acknowledged. P.C.T. Souza also acknowledges the support provided by the CNRS and PharmCADD.” No other disclosures were reported.
Figures








References
-
- Abraham, M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High-performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1:19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Berendsen, H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., and Haak J.R.. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. 10.1063/1.448118 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

