The CDK8 kinase module: A novel player in the transcription of translation initiation and ribosomal genes
- PMID: 39565680
- PMCID: PMC11742111
- DOI: 10.1091/mbc.E24-04-0164
The CDK8 kinase module: A novel player in the transcription of translation initiation and ribosomal genes
Abstract
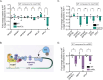
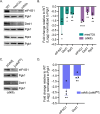
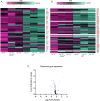
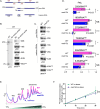
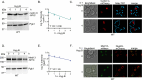
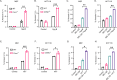
Survival following stress is dependent upon reprogramming transcription and translation. Communication between these programs following stress is critical for adaptation but is not clearly understood. The Cdk8 kinase module (CKM) of the Mediator complex modulates the transcriptional response to various stresses. Its involvement in regulating translational machinery has yet to be elucidated, highlighting an existing gap in knowledge. Here, we report that the CKM positively regulates a subset of ribosomal protein (RP) and translation initiation factor (TIF)-encoding genes under physiological conditions in Saccharomyces cerevisiae. In mouse embryonic fibroblasts and HCT116 cells, the CKM regulates unique sets of RP and TIF genes, demonstrating some conservation of function across species. In yeast, this is mediated by Cdk8 phosphorylation of one or more transcription factors which control RP and TIF expression. Conversely, the CKM is disassembled following nutrition stress, permitting repression of RP and TIF genes. The CKM also plays a transcriptional role important for promoting cell survival, particularly during translational machinery stress triggered by ribosome-targeting antibiotics. Furthermore, in mammalian cells, the activity of CDK8 and its paralogue, CDK19, promotes cell survival following ribosome inhibition. These results provide mechanistic insights into the CKM's role in regulating expression of a subset of genes associated with translation.
Figures




Similar articles
-
The Cdk8 kinase module regulates interaction of the mediator complex with RNA polymerase II.J Biol Chem. 2021 Jan-Jun;296:100734. doi: 10.1016/j.jbc.2021.100734. Epub 2021 Apr 30. J Biol Chem. 2021. PMID: 33933450 Free PMC article.
-
MED12 interacts with the heat-shock transcription factor HSF1 and recruits CDK8 to promote the heat-shock response in mammalian cells.FEBS Lett. 2021 Jul;595(14):1933-1948. doi: 10.1002/1873-3468.14139. Epub 2021 Jun 15. FEBS Lett. 2021. PMID: 34056708
-
Transcriptional Responses to IFN-γ Require Mediator Kinase-Dependent Pause Release and Mechanistically Distinct CDK8 and CDK19 Functions.Mol Cell. 2019 Nov 7;76(3):485-499.e8. doi: 10.1016/j.molcel.2019.07.034. Epub 2019 Sep 5. Mol Cell. 2019. PMID: 31495563 Free PMC article.
-
Regulatory functions of the Mediator kinases CDK8 and CDK19.Transcription. 2019 Apr;10(2):76-90. doi: 10.1080/21541264.2018.1556915. Epub 2018 Dec 26. Transcription. 2019. PMID: 30585107 Free PMC article. Review.
-
CDK8: a positive regulator of transcription.Transcription. 2010 Jul-Aug;1(1):4-12. doi: 10.4161/trns.1.1.12373. Transcription. 2010. PMID: 21327159 Free PMC article. Review.
Cited by
-
Ribosome-directed cancer therapies: the tip of the iceberg?Trends Pharmacol Sci. 2025 Apr;46(4):303-310. doi: 10.1016/j.tips.2025.02.001. Epub 2025 Mar 4. Trends Pharmacol Sci. 2025. PMID: 40044536 Review.
-
Quitting Your Day Job in Response to Stress: Cell Survival and Cell Death Require Secondary Cytoplasmic Roles of Cyclin C and Med13.Cells. 2025 Apr 25;14(9):636. doi: 10.3390/cells14090636. Cells. 2025. PMID: 40358161 Free PMC article. Review.
-
Selective Dual Inhibition of TNKS1 and CDK8 by TCS9725 Attenuates STAT1/β-Catenin/TGFβ1 Signaling in Renal Cancer.Curr Issues Mol Biol. 2025 Jun 17;47(6):463. doi: 10.3390/cimb47060463. Curr Issues Mol Biol. 2025. PMID: 40699862 Free PMC article.
References
-
- Abraham AK, Pihl A (1983). Effect of protein synthesis inhibitors on the fidelity of translation in eukaryotic systems. Biochim Biophys Acta 741, 197–203. - PubMed
-
- Adler AS, McCleland ML, Truong T, Lau S, Modrusan Z, Soukup TM, Roose-Girma M, Blackwood EM, Firestein R (2012). CDK8 maintains tumor dedifferentiation and embryonic stem cell pluripotency. Cancer Res 72, 2129–2139. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

