Genomic analysis of the early COVID-19 pandemic in Haiti reveals Caribbean-specific variant dynamics
- PMID: 39565753
- PMCID: PMC11578445
- DOI: 10.1371/journal.pgph.0003536
Genomic analysis of the early COVID-19 pandemic in Haiti reveals Caribbean-specific variant dynamics
Abstract
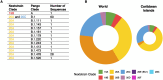
Pathogen sequencing during the COVID-19 pandemic has generated more whole genome sequencing data than for any other epidemic, allowing epidemiologists to monitor the transmission and evolution of SARS-CoV-2. However, large parts of the world are heavily underrepresented in sequencing efforts, including the Caribbean islands. We performed genome sequencing of SARS-CoV-2 from upper respiratory tract samples collected in Haiti during the spring of 2020. We used phylogenetic analysis to assess the pandemic dynamics in the Caribbean region and observed that the epidemic in Haiti was seeded by multiple introductions, primarily from the United States. We identified the emergence of a SARS-CoV-2 lineage (B.1.478) from Haiti that spread into North America, as well as evidence of the undocumented spread of SARS-CoV-2 within the Caribbean. We demonstrate that the genomic analysis of a relatively modest number of samples from a severely under-sampled region can provide new insight on a previously unobserved spread of a specific lineage, demonstrating the importance of geographically widespread genomic epidemiology.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous
