TMPRSS2 in microbial interactions: Insights from HKU1 and TcsH
- PMID: 39565778
- PMCID: PMC11578487
- DOI: 10.1371/journal.ppat.1012677
TMPRSS2 in microbial interactions: Insights from HKU1 and TcsH
Abstract
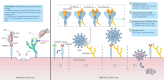
Transmembrane Serine Protease 2 (TMPRSS2), known primarily for its role as a protease, has emerged as a critical receptor for microbial agents such as human coronavirus HKU1 and exotoxin TcsH. HKU1 utilizes both sialoglycan and TMPRSS2 for cellular entry, where sialoglycan primes the spike protein for TMPRSS2 binding. TMPRSS2 undergoes autocleavage to enhance its affinity for the HKU1 spike, facilitating viral membrane fusion postcleavage. Interestingly, TMPRSS2's catalytic function is dispensable for both HKU1 and TcsH interactions, suggesting alternative roles in pathogenesis. Structural insights highlight potential therapeutic targets against viral infections and cancers, leveraging TMPRSS2 interactions for drug development. Understanding the interplay between TMPRSS2 and microbes opens new avenues for targeting TMPRSS2 in developing treatments for infections.
Copyright: © 2024 Pan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
The Transmembrane Protease Serine 2 (TMPRSS2) Non-Protease Domains Regulating Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike-Mediated Virus Entry.Viruses. 2023 Oct 19;15(10):2124. doi: 10.3390/v15102124. Viruses. 2023. PMID: 37896901 Free PMC article.
-
Structural basis of TMPRSS2 zymogen activation and recognition by the HKU1 seasonal coronavirus.Cell. 2024 Aug 8;187(16):4246-4260.e16. doi: 10.1016/j.cell.2024.06.007. Epub 2024 Jul 3. Cell. 2024. PMID: 38964326
-
TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.Cell. 2024 Aug 8;187(16):4261-4271.e17. doi: 10.1016/j.cell.2024.06.016. Epub 2024 Jul 3. Cell. 2024. PMID: 38964329
-
The discovery and development of transmembrane serine protease 2 (TMPRSS2) inhibitors as candidate drugs for the treatment of COVID-19.Expert Opin Drug Discov. 2022 Mar;17(3):231-246. doi: 10.1080/17460441.2022.2029843. Epub 2022 Jan 24. Expert Opin Drug Discov. 2022. PMID: 35072549 Free PMC article. Review.
-
Targeting the intestinal TMPRSS2 protease to prevent SARS-CoV-2 entry into enterocytes-prospects and challenges.Mol Biol Rep. 2021 May;48(5):4667-4675. doi: 10.1007/s11033-021-06390-1. Epub 2021 May 22. Mol Biol Rep. 2021. PMID: 34023987 Free PMC article. Review.
Cited by
-
TMPRSS2 as a Key Player in Viral Pathogenesis: Influenza and Coronaviruses.Biomolecules. 2025 Jan 7;15(1):75. doi: 10.3390/biom15010075. Biomolecules. 2025. PMID: 39858469 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

