Structure of the human TSC:WIPI3 lysosomal recruitment complex
- PMID: 39565846
- PMCID: PMC11578170
- DOI: 10.1126/sciadv.adr5807
Structure of the human TSC:WIPI3 lysosomal recruitment complex
Abstract
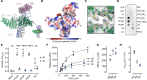
Tuberous sclerosis complex (TSC) is targeted to the lysosomal membrane, where it hydrolyzes RAS homolog-mTORC1 binding (RHEB) from its GTP-bound to GDP-bound state, inhibiting mechanistic target of rapamycin complex 1 (mTORC1). Loss-of-function mutations in TSC cause TSC disease, marked by excessive tumor growth. Here, we overcome a high degree of continuous conformational heterogeneity to determine the 2.8-Å cryo-electron microscopy (cryo-EM) structure of the complete human TSC in complex with the lysosomal recruitment factor WD repeat domain phosphoinositide-interacting protein 3 (WIPI3). We discover a previously undetected amino-terminal TSC1 HEAT repeat dimer that clamps onto a single TSC wing and forms a phosphatidylinositol phosphate (PIP)-binding pocket, which specifically binds monophosphorylated PIPs. These structural advances provide a model by which WIPI3 and PIP-signaling networks coordinate to recruit TSC to the lysosomal membrane to inhibit mTORC1. The high-resolution TSC structure reveals previously unrecognized mutational hotspots and uncovers crucial insights into the mechanisms of TSC dysregulation in disease.
Figures



References
-
- Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D., Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5, 578–581 (2003). - PubMed
-
- Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J., Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 (2003). - PubMed
-
- Castro A. F., Rebhun J. F., Clark G. J., Quilliam L. A., Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 278, 32493–32496 (2003). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

