Research note: Whole-genome sequencing revealed genomic diversity dynamics in duck conserved populations
- PMID: 39566170
- PMCID: PMC11617230
- DOI: 10.1016/j.psj.2024.104509
Research note: Whole-genome sequencing revealed genomic diversity dynamics in duck conserved populations
Abstract
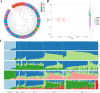
The purpose of this study was to comprehensively analyze the genetic diversity of the Pekin duck conserved population with five generations and to evaluate the effectiveness of the current conservation strategy. In total, 277 Pekin duck conserved individuals and 40 Mallards as ancestral controls were collected. Each duck was sequenced at about 10X whole-genome coverage, while over 7.4 million single nucleotide polymorphisms (SNPs) in total were detected for genetic diversity analysis. Both the expected heterozygosity and observed heterozygosity values exceeded 0.3. The genetic differentiation (FST) values ranged from 0.007 to 0.039, and the Polymorphism information content (PIC) values ranged from 0.29 to 0.34. These results indicate no significant differentiation between generations, and the genetic diversity remains high. In particular, the inbreeding coefficient has been strictly controlled and has not increased rapidly during the conservation. Overall, the inbreeding coefficient of the conserved Pekin duck population was higher than that of its wild ancestors, indicating that domestication has resulted in reduced genetic diversity. This is the first report using whole genome resequencing data to systematically evaluate the genomic dynamics across several generations in ducks. The results show that the strategy of free mating and random seed retention within sire families is effective for maintaining the genetic diversity of the conserved Pekin duck population.
Keywords: Conservation strategy; Conserved population; Duck; Genetic diversity.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could appear to influence the work reported in this paper.
Figures

References
-
- Boettcher P.J., Tixier-Boichard M., Toro M.A., Simianer H., Eding H., Gandini G., Joost S., Garcia D., Colli L., Ajmone-Marsan P., Consortium G. Objectives, criteria and methods for using molecular genetic data in priority setting for conservation of animal genetic resources. Anim. Genet. 2010;1:64–77. doi: 10.1111/j.1365-2052.2010.02050.x. - DOI - PubMed
-
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., Genomes Project Analysis G. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

