Solid tumor immunotherapy using NKG2D-based adaptor CAR T cells
- PMID: 39566469
- PMCID: PMC11604534
- DOI: 10.1016/j.xcrm.2024.101827
Solid tumor immunotherapy using NKG2D-based adaptor CAR T cells
Abstract
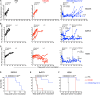
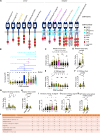
NKG2D ligands (NKG2DLs) are broadly expressed in cancer. To target these, we describe an adaptor chimeric antigen receptor (CAR) termed NKG2D/Dap10-12. Herein, T cells are engineered to co-express NKG2D with a fusion protein that comprises Dap10 joined to a Dap12 endodomain. NKG2D/Dap10-12 T cells elicit compelling efficacy, eradicating or controlling NKG2DL-expressing tumors in several established xenograft models. Importantly, durable responses, long-term survival, and rejection of tumor re-challenge are reproducibly achieved. Efficacy is markedly superior to a clinical stage CAR analog, comprising an NKG2D-CD3ζ fusion. Structure-function analysis using an extended CAR panel demonstrates that potency is dependent on membrane proximity of signaling units, high NKG2D cell surface expression, adaptor structure, provision of exogenous Dap10, and inclusion of one rather than three immune tyrosine activation motifs per signaling unit. Potent therapeutic impact of NKG2D/Dap10-12 T cells is also underpinned by enhanced oxidative phosphorylation, reduced senescence, and transcriptomic re-programming for increased ribosomal biogenesis.
Keywords: Dap10; Dap12; NKG2D; adaptor; chimeric antigen receptor.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.M. is CSO, scientific founder, and shareholder of Leucid Bio. M.G., F.K., C.M.H., R.M., C.B., C.T., and D.M.D. are employees of Leucid Bio while P.D. is a former Leucid Bio employee. D.L.-Y. is undertaking a PhD studentship funded by Leucid Bio and has acted as a consultant for Leucid Bio. J.M., D.M.D., D.L.-Y., and F.K. are co-inventors on patent filings in relation to adaptor CAR technology.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

