Leveraging the power of long reads for targeted sequencing
- PMID: 39567237
- PMCID: PMC11610587
- DOI: 10.1101/gr.279168.124
Leveraging the power of long reads for targeted sequencing
Abstract
Long-read sequencing technologies have improved the contiguity and, as a result, the quality of genome assemblies by generating reads long enough to span and resolve complex or repetitive regions of the genome. Several groups have shown the power of long reads in detecting thousands of genomic and epigenomic features that were previously missed by short-read sequencing approaches. While these studies demonstrate how long reads can help resolve repetitive and complex regions of the genome, they also highlight the throughput and coverage requirements needed to accurately resolve variant alleles across large populations using these platforms. At the time of this review, whole-genome long-read sequencing is more expensive than short-read sequencing on the highest throughput short-read instruments; thus, achieving sufficient coverage to detect low-frequency variants (such as somatic variation) in heterogenous samples remains challenging. Targeted sequencing, on the other hand, provides the depth necessary to detect these low-frequency variants in heterogeneous populations. Here, we review currently used and recently developed targeted sequencing strategies that leverage existing long-read technologies to increase the resolution with which we can look at nucleic acids in a variety of biological contexts.
© 2024 Iyer et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
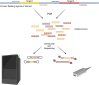

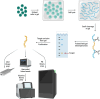
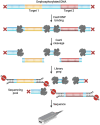
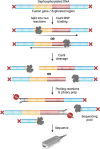

Similar articles
-
Unraveling the hidden complexity of cancer through long-read sequencing.Genome Res. 2025 Apr 14;35(4):599-620. doi: 10.1101/gr.280041.124. Genome Res. 2025. PMID: 40113261 Free PMC article. Review.
-
Long-read human genome sequencing and its applications.Nat Rev Genet. 2020 Oct;21(10):597-614. doi: 10.1038/s41576-020-0236-x. Epub 2020 Jun 5. Nat Rev Genet. 2020. PMID: 32504078 Free PMC article. Review.
-
Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing.BMC Genomics. 2019 Jan 9;20(1):23. doi: 10.1186/s12864-018-5381-7. BMC Genomics. 2019. PMID: 30626323 Free PMC article.
-
The impact of long-read sequencing on human population-scale genomics.Genome Res. 2025 Apr 14;35(4):593-598. doi: 10.1101/gr.280120.124. Genome Res. 2025. PMID: 40228902 Review.
-
Improvements in Genomic Technologies: Application to Crop Genomics.Trends Biotechnol. 2017 Jun;35(6):547-558. doi: 10.1016/j.tibtech.2017.02.009. Epub 2017 Mar 9. Trends Biotechnol. 2017. PMID: 28284542 Review.
Cited by
-
Long-read sequencing for diagnosis of genetic myopathies.BMJ Neurol Open. 2025 May 11;7(1):e000990. doi: 10.1136/bmjno-2024-000990. eCollection 2025. BMJ Neurol Open. 2025. PMID: 40357124 Free PMC article. Review.
-
Unraveling the hidden complexity of cancer through long-read sequencing.Genome Res. 2025 Apr 14;35(4):599-620. doi: 10.1101/gr.280041.124. Genome Res. 2025. PMID: 40113261 Free PMC article. Review.
-
Early Genetic Evolution of Driver Mutations in Uveal Melanoma.medRxiv [Preprint]. 2025 Jun 12:2025.06.10.25329358. doi: 10.1101/2025.06.10.25329358. medRxiv. 2025. PMID: 40585173 Free PMC article. Preprint.
-
Targeted long-read sequencing to quantify methylation of the C9orf72 repeat expansion.Mol Neurodegener. 2024 Dec 21;19(1):99. doi: 10.1186/s13024-024-00790-0. Mol Neurodegener. 2024. PMID: 39709476 Free PMC article.
-
Long-Read Sequencing and Structural Variant Detection: Unlocking the Hidden Genome in Rare Genetic Disorders.Diagnostics (Basel). 2025 Jul 17;15(14):1803. doi: 10.3390/diagnostics15141803. Diagnostics (Basel). 2025. PMID: 40722552 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
