Identification of pancreatic cancer-specific protease substrates for protease-dependent targeted delivery
- PMID: 39567504
- PMCID: PMC11579016
- DOI: 10.1038/s41389-024-00542-1
Identification of pancreatic cancer-specific protease substrates for protease-dependent targeted delivery
Abstract
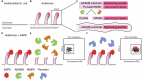
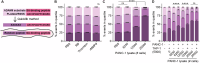
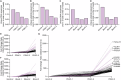
Pancreatic ductal adenocarcinoma (PDAC) presents significant challenges due to the inadequacy of existing chemotherapeutics, which often result in toxicity-dependent dose limitations and premature cessation of therapy. Targeted delivery of therapeutic molecules offers a promising solution. Given that PDAC is marked by a desmoplastic reaction with extensive aberrant protease activity, protease-dependent targeted delivery could minimize off-target toxicities and is of increasing interest. The efficacy of targeted delivery hinges on the specificity of the substrates used; insufficient specificity can lead to off-target effects, reducing the advantage over non-targeted methods. Here, we employ an unbiased library approach to screen over 7 million peptide substrates for proteolytic cleavage by PDAC cell lysates, identifying 37 substrates enriched by at least 500-fold after three rounds of selection. As systemically administered targeted delivery depends on the absence of substrate cleavage in circulation, the peptide library was also screened against whole blood lysates, and enriched substrates were removed from the PDAC-enriched dataset to obtain PDAC-specific substrates. In vitro validation using FRET-peptides showed that 13 of the selected 15 substrates are cleaved by a panel of PDAC cell line lysates. Moreover, evaluation against healthy murine organ and human blood lysates to assess off-target cleavage revealed that the identified substrates are indeed PDAC-specific and that several substrates may be superior with respect to PDAC specificity over the CAPN2-responsive substrate, which has recently shown preclinical potential in targeted therapy, but future animal models should address the potential superiority. Overall, we thus identified substrates with high selectivity and sensitivity for PDAC that could be employed in protease-dependent targeted therapies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: M.F.B. has received research funding from Celgene, Lead Pharma, and Frame Therapeutics, and acted as a consultant to Servier, Olympus, and Wholomics. None of these were involved in the design of this study nor the drafting of the paper. Ethical approval: The use of whole blood was evaluated and approved by the Ethics Committee of the Amsterdam University Medical Centers and performed following the instructions from the Declaration of Helsinki. The animal tissues were collected in accordance with the regulations on animal experimentation and approved by the ethics committee of Amsterdam University Medical Centers (Approval number: AVD118002015222). Ethics approval and consent to participate: All methods were performed in accordance with the relevant guidelines and regulations. Informed consent: The blood donor has provided consent for the usage of the collected blood in scientific research.
Figures
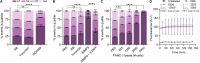

Similar articles
-
CAPN2-responsive mesoporous silica nanoparticles: A promising nanocarrier for targeted therapy of pancreatic cancer.Cancer Lett. 2024 May 28;590:216845. doi: 10.1016/j.canlet.2024.216845. Epub 2024 Apr 6. Cancer Lett. 2024. PMID: 38589004
-
ADAM9-Responsive Mesoporous Silica Nanoparticles for Targeted Drug Delivery in Pancreatic Cancer.Cancers (Basel). 2021 Jul 1;13(13):3321. doi: 10.3390/cancers13133321. Cancers (Basel). 2021. PMID: 34282781 Free PMC article.
-
Preclinical Assessment of ADAM9-Responsive Mesoporous Silica Nanoparticles for the Treatment of Pancreatic Cancer.Int J Mol Sci. 2023 Jun 27;24(13):10704. doi: 10.3390/ijms241310704. Int J Mol Sci. 2023. PMID: 37445886 Free PMC article.
-
Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma.J Hematol Oncol. 2020 Oct 2;13(1):130. doi: 10.1186/s13045-020-00958-3. J Hematol Oncol. 2020. PMID: 33008426 Free PMC article. Review.
-
Targeted Therapies for Pancreatic Cancer and Hurdles Ahead.Anticancer Res. 2018 Dec;38(12):6591-6606. doi: 10.21873/anticanres.13026. Anticancer Res. 2018. PMID: 30504367 Review.
References
-
- Kappelhoff R, Puente XS, Wilson CH, Seth A, López-Otín C, Overall CM. Overview of transcriptomic analysis of all human proteases, non-proteolytic homologs and inhibitors: organ, tissue and ovarian cancer cell line expression profiling of the human protease degradome by the CLIP-CHIP™ DNA microarray. Biochim Biophys Acta Mol Cell Res. 2017;1864:2210–9. - PubMed
-
- Doucet A, Butler GS, Rodríguez D, Prudova A, Overall CM. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteom. 2008;7:1925–51. - PubMed
-
- Walsh PN, Ahmad SS. Proteases in blood clotting. Essays Biochem. 2002;38:95–111. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

