Disorganized chromatin hierarchy and stem cell aging in a male patient of atypical laminopathy-based progeria mandibuloacral dysplasia type A
- PMID: 39567511
- PMCID: PMC11579472
- DOI: 10.1038/s41467-024-54338-3
Disorganized chromatin hierarchy and stem cell aging in a male patient of atypical laminopathy-based progeria mandibuloacral dysplasia type A
Abstract
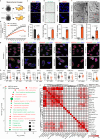
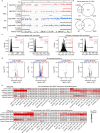
Studies of laminopathy-based progeria offer insights into aging-associated diseases and highlight the role of LMNA in chromatin organization. Mandibuloacral dysplasia type A (MAD) is a largely unexplored form of atypical progeria that lacks lamin A post-translational processing defects. Using iPSCs derived from a male MAD patient carrying homozygous LMNA p.R527C, premature aging phenotypes are recapitulated in multiple mesenchymal lineages, including mesenchymal stem cells (MSCs). Comparison with 26 human aging MSC expression datasets reveals that MAD-MSCs exhibit the highest similarity to senescent primary human MSCs. Lamina-chromatin interaction analysis reveals reorganization of lamina-associating domains (LADs) and repositioning of non-LAD binding peaks may contribute to the observed accelerated senescence. Additionally, 3D genome organization further supports hierarchical chromatin disorganization in MAD stem cells, alongside dysregulation of genes involved in epigenetic modification, stem cell fate maintenance, senescence, and geroprotection. Together, these findings suggest LMNA missense mutation is linked to chromatin alterations in an atypical progeroid syndrome.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

