Staphylococcus epidermidis alters macrophage polarization and phagocytic uptake by extracellular DNA release in vitro
- PMID: 39567551
- PMCID: PMC11579364
- DOI: 10.1038/s41522-024-00604-7
Staphylococcus epidermidis alters macrophage polarization and phagocytic uptake by extracellular DNA release in vitro
Abstract
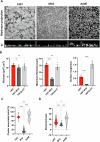
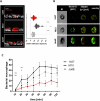
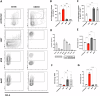
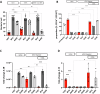
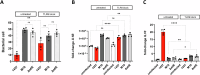
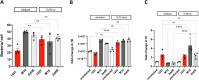
Biofilm formation shields Staphylococcus epidermidis from host defense mechanisms, contributing to chronic implant infections. Using wild-type S. epidermidis 1457, a PIA-negative mutant (1457-M10), and an eDNA-negative mutant (1457ΔatlE), this study examined the influence of biofilm matrix components on human monocyte-derived macrophage (hMDM) interactions. The wild-type strain was resistant to phagocytosis and induced an anti-inflammatory response in hMDMs, while both mutants were more susceptible to phagocytosis and triggered a pro-inflammatory response. Removing eDNA from the 1457 biofilm matrix increased hMDM uptake and a pro-inflammatory reaction, whereas adding eDNA to the 1457ΔatlE mutant reduced phagocytosis and promoted an anti-inflammatory response. Inhibiting TLR9 enhanced bacterial uptake and induced a pro-inflammatory response in hMDMs exposed to wild-type S. epidermidis. This study highlights the critical role of eDNA in immune evasion and the central role of TLR9 in modulating macrophage responses, advancing the understanding of implant infections.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Rosenstein, R. & Gotz, F. What distinguishes highly pathogenic staphylococci from medium- and non-pathogenic? Curr. Top. Microbiol. Immunol.358, 33–89 (2013). - PubMed
-
- Ali, N., Adil, S. N. & Shaikh, M. U. Bloodstream and central line isolates from hematopoietic stem cell transplant recipients: data from a developing country. Transpl. Infect. Dis.16, 98–105 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

