Broadly inhibitory antibodies to severe malaria virulence proteins
- PMID: 39567685
- PMCID: PMC12338075
- DOI: 10.1038/s41586-024-08220-3
Broadly inhibitory antibodies to severe malaria virulence proteins
Abstract
Malaria pathology is driven by the accumulation of Plasmodium falciparum-infected erythrocytes in microvessels1. This process is mediated by the polymorphic erythrocyte membrane protein 1 (PfEMP1) adhesion proteins of the parasite. A subset of PfEMP1 variants that bind to human endothelial protein C receptor (EPCR) through their CIDRα1 domains is responsible for severe malaria pathogenesis2. A longstanding question is whether individual antibodies can recognize the large repertoire of circulating PfEMP1 variants. Here we describe two broadly reactive and inhibitory human monoclonal antibodies to CIDRα1. The antibodies isolated from two different individuals exhibited similar and consistent EPCR-binding inhibition of diverse CIDRα1 domains, representing five of the six subclasses of CIDRα1. Both antibodies inhibited EPCR binding of both recombinant full-length and native PfEMP1 proteins, as well as parasite sequestration in bioengineered 3D human brain microvessels under physiologically relevant flow conditions. Structural analyses of the two antibodies in complex with three different CIDRα1 antigen variants reveal similar binding mechanisms that depend on interactions with three highly conserved amino acid residues of the EPCR-binding site in CIDRα1. These broadly reactive antibodies are likely to represent a common mechanism of acquired immunity to severe malaria and offer novel insights for the design of a vaccine or treatment targeting severe malaria.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











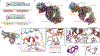
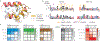
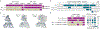
Update of
-
Broadly inhibitory antibodies against severe malaria virulence proteins.bioRxiv [Preprint]. 2024 Jan 25:2024.01.25.577124. doi: 10.1101/2024.01.25.577124. bioRxiv. 2024. Update in: Nature. 2024 Dec;636(8041):182-189. doi: 10.1038/s41586-024-08220-3. PMID: 38328068 Free PMC article. Updated. Preprint.
References
-
- WHO. World malaria report 2023. World Health Organization (2023).
-
- Baruch DI et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82, 77–87 (1995). https://doi.org/0092-8674(95)90054-3 [pii] - PubMed
-
- Smith JD et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110 (1995). https://doi.org/0092-8674(95)90056-X [pii] - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

