Liver X receptor unlinks intestinal regeneration and tumorigenesis
- PMID: 39567700
- PMCID: PMC11779645
- DOI: 10.1038/s41586-024-08247-6
Liver X receptor unlinks intestinal regeneration and tumorigenesis
Abstract
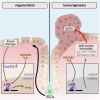
Uncontrolled regeneration leads to neoplastic transformation1-3. The intestinal epithelium requires precise regulation during continuous homeostatic and damage-induced tissue renewal to prevent neoplastic transformation, suggesting that pathways unlinking tumour growth from regenerative processes must exist. Here, by mining RNA-sequencing datasets from two intestinal damage models4,5 and using pharmacological, transcriptomics and genetic tools, we identified liver X receptor (LXR) pathway activation as a tissue adaptation to damage that reciprocally regulates intestinal regeneration and tumorigenesis. Using single-cell RNA sequencing, intestinal organoids, and gain- and loss-of-function experiments, we demonstrate that LXR activation in intestinal epithelial cells induces amphiregulin (Areg), enhancing regenerative responses. This response is coordinated by the LXR-ligand-producing enzyme CYP27A1, which was upregulated in damaged intestinal crypt niches. Deletion of Cyp27a1 impaired intestinal regeneration, which was rescued by exogenous LXR agonists. Notably, in tumour models, Cyp27a1 deficiency led to increased tumour growth, whereas LXR activation elicited anti-tumour responses dependent on adaptive immunity. Consistently, human colorectal cancer specimens exhibited reduced levels of CYP27A1, LXR target genes, and B and CD8 T cell gene signatures. We therefore identify an epithelial adaptation mechanism to damage, whereby LXR functions as a rheostat, promoting tissue repair while limiting tumorigenesis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: E.J.V. has received research grants from F. Hoffmann-La Roche and is a founder of PaperVids. S.D. works as a consultant for Cellphi Biotechnology. The other authors declare no competing interests.
Figures














References
-
- Oudhoff, M. J. et al. SETD7 controls intestinal regeneration and tumorigenesis by regulating Wnt/β-catenin and Hippo/YAP signaling. Dev. Cell37, 47–57 (2016). - PubMed
-
- Ayyaz, A. et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature569, 121–125 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

