Spatially resolved single-cell atlas unveils a distinct cellular signature of fatal lung COVID-19 in a Malawian population
- PMID: 39567718
- PMCID: PMC11645280
- DOI: 10.1038/s41591-024-03354-3
Spatially resolved single-cell atlas unveils a distinct cellular signature of fatal lung COVID-19 in a Malawian population
Abstract
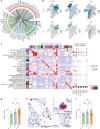
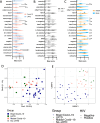
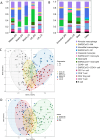
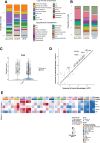
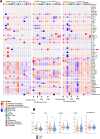
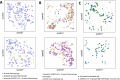
Postmortem single-cell studies have transformed understanding of lower respiratory tract diseases (LRTDs), including coronavirus disease 2019 (COVID-19), but there are minimal data from African settings where HIV, malaria and other environmental exposures may affect disease pathobiology and treatment targets. In this study, we used histology and high-dimensional imaging to characterize fatal lung disease in Malawian adults with (n = 9) and without (n = 7) COVID-19, and we generated single-cell transcriptomics data from lung, blood and nasal cells. Data integration with other cohorts showed a conserved COVID-19 histopathological signature, driven by contrasting immune and inflammatory mechanisms: in US, European and Asian cohorts, by type I/III interferon (IFN) responses, particularly in blood-derived monocytes, and in the Malawian cohort, by response to IFN-γ in lung-resident macrophages. HIV status had minimal impact on histology or immunopathology. Our study provides a data resource and highlights the importance of studying the cellular mechanisms of disease in underrepresented populations, indicating shared and distinct targets for treatment.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









References
-
- Lindeboom, R. G. H., Regev, A. & Teichmann, S. A. Towards a Human Cell Atlas: taking notes from the past. Trends Genet.37, 625–630 (2021). - PubMed
-
- Rood, J. E., Maartens, A., Hupalowska, A., Teichmann, S. A. & Regev, A. Impact of the Human Cell Atlas on medicine. Nat. Med.28, 2486–2496 (2022). - PubMed
-
- Majumder, P. P., Mhlanga, M. M. & Shalek, A. K. The Human Cell Atlas and equity: lessons learned. Nat. Med.26, 1509–1511 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

