Consensus prediction of cell type labels in single-cell data with popV
- PMID: 39567746
- PMCID: PMC11631762
- DOI: 10.1038/s41588-024-01993-3
Consensus prediction of cell type labels in single-cell data with popV
Abstract
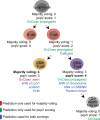
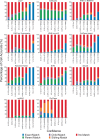
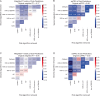
Cell-type classification is a crucial step in single-cell sequencing analysis. Various methods have been proposed for transferring a cell-type label from an annotated reference atlas to unannotated query datasets. Existing methods for transferring cell-type labels lack proper uncertainty estimation for the resulting annotations, limiting interpretability and usefulness. To address this, we propose popular Vote (popV), an ensemble of prediction models with an ontology-based voting scheme. PopV achieves accurate cell-type labeling and provides uncertainty scores. In multiple case studies, popV confidently annotates the majority of cells while highlighting cell populations that are challenging to annotate by label transfer. This additional step helps to reduce the load of manual inspection, which is often a necessary component of the annotation process, and enables one to focus on the most problematic parts of the annotation, streamlining the overall annotation process.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









References
-
- Clarke, Z. A. et al. Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nat. Protoc.16, 2749–2764 (2021). - PubMed

