Luminal breast epithelial cells of BRCA1 or BRCA2 mutation carriers and noncarriers harbor common breast cancer copy number alterations
- PMID: 39567747
- PMCID: PMC11631757
- DOI: 10.1038/s41588-024-01988-0
Luminal breast epithelial cells of BRCA1 or BRCA2 mutation carriers and noncarriers harbor common breast cancer copy number alterations
Abstract
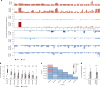
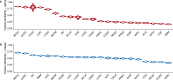
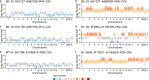
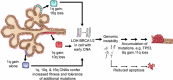
The prevalence and nature of somatic copy number alterations (CNAs) in breast epithelium and their role in tumor initiation and evolution remain poorly understood. Using single-cell DNA sequencing (49,238 cells) of epithelium from BRCA1 and BRCA2 carriers or wild-type individuals, we identified recurrent CNAs (for example, 1q-gain and 7q, 10q, 16q and 22q-loss) that are present in a rare population of cells across almost all samples (n = 28). In BRCA1/BRCA2 carriers, these occur before loss of heterozygosity (LOH) of wild-type alleles. These CNAs, common in malignant tumors, are enriched in luminal cells but absent in basal myoepithelial cells. Allele-specific analysis of prevalent CNAs reveals that they arose by independent mutational events, consistent with convergent evolution. BRCA1/BRCA2 carriers contained a small percentage of cells with extreme aneuploidy, featuring loss of TP53, BRCA1/BRCA2 LOH and multiple breast cancer-associated CNAs. Our findings suggest that CNAs arising in normal luminal breast epithelium are precursors to clonally expanded tumor genomes.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: J.S.B. is a scientific advisory board (SAB) member of Frontier Medicines. D.A.D. is on the SAB for Oncology Analytics, has consulted for Novartis and receives research support from Canon. J.E.G. is a paid consultant for Helix and an uncompensated consultant for Konica Minolta and Earli. S.P.S. has consulted for AstraZeneca and has received funding from Bristol Meyers Squibb. S. Aparicio is cofounder and shareholder of Genome Therapeutics, uncompensated advisor to Chordia Therapeutics and advisor to Sangamo Therapeutics. The other authors declare no competing interests.
Figures









Update of
-
Luminal breast epithelial cells from wildtype and BRCA mutation carriers harbor copy number alterations commonly associated with breast cancer.bioRxiv [Preprint]. 2024 May 3:2024.05.01.591587. doi: 10.1101/2024.05.01.591587. bioRxiv. 2024. Update in: Nat Genet. 2024 Dec;56(12):2753-2762. doi: 10.1038/s41588-024-01988-0. PMID: 38746396 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

