FNDC1 is a myokine that promotes myogenesis and muscle regeneration
- PMID: 39567831
- PMCID: PMC11695938
- DOI: 10.1038/s44318-024-00285-0
FNDC1 is a myokine that promotes myogenesis and muscle regeneration
Abstract
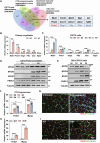
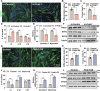
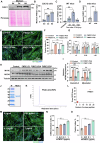
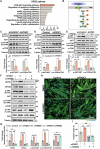
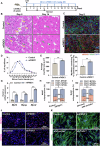
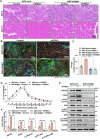
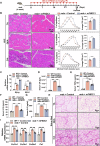
Myogenesis is essential for skeletal muscle formation and regeneration after injury, yet its regulators are largely unknown. Here we identified fibronectin type III domain containing 1 (FNDC1) as a previously uncharacterized myokine. In vitro studies showed that knockdown of Fndc1 in myoblasts reduces myotube formation, while overexpression of Fndc1 promotes myogenic differentiation. We further generated recombinant truncated mouse FNDC1 (mFNDC1), which retains reliable activity in promoting myoblast differentiation in vitro. Gain- and loss-of-function studies collectively showed that FNDC1 promotes cardiotoxin (CTX)-induced muscle regeneration in adult mice. Furthermore, recombinant FNDC1 treatment ameliorated pathological muscle phenotypes in the mdx mouse model of Duchenne muscular dystrophy. Mechanistically, FNDC1 bound to the integrin α5β1 and activated the downstream FAK/PI3K/AKT/mTOR pathway to promote myogenic differentiation. Pharmacological inhibition of integrin α5β1 or of the downstream FAK/PI3K/AKT/mTOR pathway abolished the pro-myogenic effect of FNDC1. Collectively, these results suggested that myokine FNDC1 might be used as a therapeutic agent to regulate myogenic differentiation and muscle regeneration for the treatment of acute and chronic muscle disease.
Keywords: FNDC1; Integrin α5β1; Muscle Regeneration; Myogenic Differentiation.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures


References
-
- Boppart MD, Burkin DJ, Kaufman SJ (2006) Alpha7beta1-integrin regulates mechanotransduction and prevents skeletal muscle injury. Am J Physiol Cell Physiol 290:C1660–C1665 - PubMed
MeSH terms
Substances
Grants and funding
- 2021YFF1000602/MOST | National Key Research and Development Program of China (NKPs)
- 2023ZD0404702/STI2030 Major Projects
- 2023-CX-TD-57/| Key Science and Technology Program of Shaanxi Province ( )
- 2023GXJS-02-01/Shaanxi Livestock and Poultry Breeding Common Technology Research and Development Platform
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

