Identification, genomic localization, and functional validation of salt-stress-related lncRNAs in Indian Mustard (Brassica juncea L.)
- PMID: 39567864
- PMCID: PMC11580500
- DOI: 10.1186/s12864-024-10964-1
Identification, genomic localization, and functional validation of salt-stress-related lncRNAs in Indian Mustard (Brassica juncea L.)
Abstract
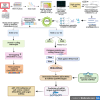
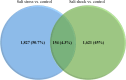
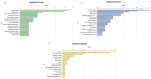
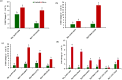
Indian Mustard (Brassica juncea L.) is a globally cultivated winter oilseed crop of the rapeseed-mustard group. It is predominantly grown in the semi-arid northwest agroclimatic zone of India, characterized by high soil salinity. Enhancing tolerance to salt stress in B. juncea is therefore crucial for sustaining its production in this region. Long non-coding RNAs (lncRNAs) play critical roles in coordinating gene expression under various abiotic stresses, including salt stress, but their involvement in the salt stress response in B. juncea remains largely unknown. In this study, we conducted RNA-seq analysis on control, salt-stressed, and salt-shocked young leaves of the salt-tolerant B. juncea cv CS-52. We identified a total of 3,602 differentially expressed transcripts between stress versus control and shock versus control samples. Among these, 61 were identified as potential lncRNAs, with 21 specific to salt stress and 40 specific to salt shock. Of the 21 lncRNAs specific to salt stress, 15 were upregulated and six were downregulated, while all 40 lncRNAs unique to salt shock were downregulated. Chromosomal distribution analysis of the lncRNAs revealed their uneven placement across 18 chromosomes in B. juncea. RNA-RNA interaction analysis between salt stress-upregulated lncRNAs and salt stress-related miRNAs identified 26 interactions between 10 lncRNAs and 23 miRNAs and predicted 13 interactions between six miRNAs and 13 mRNAs. Finally, six lncRNA-miRNA-mRNA interaction networks were established, involving five lncRNAs, 13 miRNAs, and 23 mRNAs. RT-qPCR analysis revealed the upregulation of four out of five lncRNAs, along with their target mRNAs, supporting their involvement in the salt stress response in B. juncea.
Keywords: Brassica juncea; Salt shock; Salt stress; eTMs; lncRNA.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was conducted following all ethical guidelines and principles and was approved by the competent authority of the Institute. All participants provided informed consent before participating in the study. Consent for publication: All authors have agreed to the publication of this manuscript. The authors declare that they have no competing interests. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
MYB transcription factors, their regulation and interactions with non-coding RNAs during drought stress in Brassica juncea.BMC Plant Biol. 2024 Oct 24;24(1):999. doi: 10.1186/s12870-024-05736-8. BMC Plant Biol. 2024. PMID: 39448923 Free PMC article.
-
Identification of lncRNAs regulating seed traits in Brassica juncea and development of a comprehensive seed omics database.Funct Integr Genomics. 2024 Oct 15;24(5):189. doi: 10.1007/s10142-024-01470-4. Funct Integr Genomics. 2024. PMID: 39404887
-
Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa.BMC Genomics. 2019 Mar 19;20(1):227. doi: 10.1186/s12864-019-5593-5. BMC Genomics. 2019. PMID: 30890148 Free PMC article.
-
Integrated analysis of long non-coding RNAs and mRNAs reveals the regulatory network of maize seedling root responding to salt stress.BMC Genomics. 2022 Jan 13;23(1):50. doi: 10.1186/s12864-021-08286-7. BMC Genomics. 2022. PMID: 35026983 Free PMC article.
-
Current achievements and future prospects of genetic engineering in Indian mustard (Brassica juncea L. Czern & Coss.).Planta. 2020 Sep 20;252(4):56. doi: 10.1007/s00425-020-03461-8. Planta. 2020. PMID: 32951089 Review.
References
-
- Shavrukov Y. Salt stress or salt shock: which genes are we studying? J Exp Bot. 2013;64(1):119–27. 10.1093/jxb/ers316. - PubMed
-
- Mazhar S, Pellegrini E, Contin M, Bravo C, De Nobili M. Impacts of salinization caused by sea level rise on the biological processes of coastal soils - a review. Front Environ Sci. 2022;10. 10.3389/fenvs.2022.909415.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

