Detection of diagnostic somatic copy number alterations from cerebrospinal fluid cell-free DNA in brain tumor patients
- PMID: 39568088
- PMCID: PMC11580493
- DOI: 10.1186/s40478-024-01887-9
Detection of diagnostic somatic copy number alterations from cerebrospinal fluid cell-free DNA in brain tumor patients
Abstract
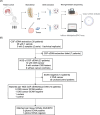
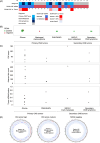
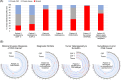
The gold standard for precise diagnostic classification of brain tumors requires tissue sampling, which carries relevant procedural risks. Brain biopsies often have limited sensitivity and fail to address tumor heterogeneity, because small tissue parts are being examined. This study aims to explore the detection and quantification of diagnostically relevant somatic copy number aberrations (SCNAs) in cell-free DNA (cfDNA) extracted from cerebrospinal fluid (CSF) in a real-world cohort of patients with defined brain tumor subtypes. A total of 33 CSF samples were collected from 30 patients for cfDNA extraction. Shallow whole-genome sequencing was conducted on CSF samples containing > 3ng of cfDNA and corresponding tissue DNA from nine patients. The sequencing cohort encompassed 26 samples of 23 patients, comprising 12 with confirmed CNS cancer as compared to 11 patients with either ambiguous CNS lesions (n = 5) or non-cancer CNS lesions (n = 6). After mapping and quality filtering SCNAs were called by depth-of-coverage analyses with a binning of 5.5 Mbp. SCNAs were exclusively identified in CSF cfDNA from brain tumor patients (10/12, 83%). In tumor patients, SCNAs were detectable in cfDNA from all patients with, but also in five of seven patients without tumor cells detected by CSF cytopathology. A substantial number of shared SCNAs were traceable between tissue and CSF in matched pair analyses. Additionally, some SCNAs unique to either CSF or tissue indicating spatial heterogeneity or tumor evolution. Also, diagnostically relevant genomic alterations as well as essential and desirable SCNAs as implemented in the current WHO classification of CNS tumors for certain primary brain tumor subtypes were traceable. In summary, this minimally invasive cfDNA-based LB approach employing shallow whole genome sequencing demonstrates potential for providing a molecularly informed diagnosis of CNS cancers, mapping tumor heterogeneity, tracking tumor evolution, and surveilling tumor patients. Further prospective trials are warranted.
Keywords: Brain tumor; Cell-free DNA; Cerebrospinal fluid; Liquid biopsy; Next-generation sequencing.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This retrospective investigation and use of patient material and data was approved by the Ethics Committee of the University Hospital Frankfurt (SNO-9-2022). Consent for publication: Not applicable. Competing interests: NGS was conducted and analyzed at Chronix Biomedical by JB, KBK and ES, who are employees of Chronix Biomedical an Oncocyte Company. JPS has received honoraria for lectures, advisory board participation, consulting, and travel grants from Abbvie, Roche, Boehringer, Bristol-Myers Squibb, Medac, Mundipharma, and UCB. PJW has received consulting fees and honoraria for lectures by Bayer, Sanofi, Janssen-Cilag, Novartis, Roche, MSD, Astellas Pharma, Bristol-Myers Squibb, Thermo Fisher Scientific, Molecular Health, Guardant Health, Sophia Genetics, Qiagen, Eli Lilly, Myriad, Hedera Dx, and Astra Zeneca and research support was provided by Astra Zeneca and Roche. HR received honoraria from AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Chop GmbH, Diaceutics, GlaxoSmithKline, HUeG, Janssen-Cilag, MCI, Merck, Novartis, Roche Pharma, Sanofi and Wolfsburg Klinikum and funding from Bristol-Myers Squibb. MWR has received a research grant from UCB. PSZ has received a lecture honorarium from Bristol-Myers Squibb. The remaining authors declare that the research was carried out without any commercial or financial relationships that could potentially create a conflict of interest.
Figures

Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Ultra-low-input cell-free DNA sequencing for tumor detection and characterization in a real-world pediatric brain tumor cohort.Acta Neuropathol Commun. 2025 Jun 28;13(1):134. doi: 10.1186/s40478-025-02024-w. Acta Neuropathol Commun. 2025. PMID: 40579709 Free PMC article.
-
High detection rate of circulating-tumor DNA from cerebrospinal fluid of children with central nervous system germ cell tumors.Acta Neuropathol Commun. 2024 Nov 20;12(1):178. doi: 10.1186/s40478-024-01886-w. Acta Neuropathol Commun. 2024. PMID: 39568077 Free PMC article.
-
CSF tau and the CSF tau/ABeta ratio for the diagnosis of Alzheimer's disease dementia and other dementias in people with mild cognitive impairment (MCI).Cochrane Database Syst Rev. 2017 Mar 22;3(3):CD010803. doi: 10.1002/14651858.CD010803.pub2. Cochrane Database Syst Rev. 2017. PMID: 28328043 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
Cited by
-
Liquid biopsies for the monitoring of gliomas and brain metastases in adults.Acta Neuropathol. 2025 Apr 26;149(1):37. doi: 10.1007/s00401-025-02880-9. Acta Neuropathol. 2025. PMID: 40285800 Free PMC article. Review.
-
Cerebrospinal Fluid-Derived Genomic Alterations Tracking Glioma.J Mol Neurosci. 2025 Jun 20;75(3):79. doi: 10.1007/s12031-025-02361-4. J Mol Neurosci. 2025. PMID: 40537713
References
-
- Seoane J, De Mattos-Arruda L, Le RE, Bardelli A, Weller M (2019) Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann Oncol 30:211–8 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

