A polygenic risk score model for psoriasis based on the protein interactions of psoriasis susceptibility loci
- PMID: 39568675
- PMCID: PMC11576467
- DOI: 10.3389/fgene.2024.1451679
A polygenic risk score model for psoriasis based on the protein interactions of psoriasis susceptibility loci
Abstract
Introduction: Polygenic Risk Scores (PRS) are an emerging tool for predicting an individual's genetic risk to a complex trait. Several methods have been proposed to construct and calculate these scores. Here, we develop a biologically driven PRS using the UK BioBank cohort through validated protein interactions (PPI) and network construction for psoriasis, incorporating variants mapped to the interacting genes of 14 psoriasis susceptibility (PSORS) loci, as identified from previous genetic linkage studies.
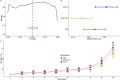
Methods: We constructed the PPI network via the implementation of two major meta-databases of protein interactions, and identified variants mapped to the identified PSORS-interacting genes. We selected only European unrelated participants including individuals with psoriasis and randomly selected healthy controls using an at least 1:4 ratio to maximize statistical power. We next compared our PPI-PRS model to (i) clinical risk models and (ii) conventional PRS calculations through p-value thresholding.
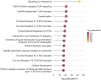
Results: Our PPI-PRS model provides comparable results to both clinical risk models and conventional approaches, despite the incorporation of a limited number of variants which have not necessarily reached genome-wide significance (GWS). Exclusion of variants mapped to the HLA-C locus, an established risk locus for psoriasis resulted in highly similar associations compared to our primary model, indicating the contribution of the genetic variability mapped to non-GWS variants in PRS computations.
Discussion: Our findings support the implementation of biologically driven approaches in PRS calculations in psoriasis, highlighting their potential clinical utility in risk assessment and treatment management.
Keywords: genome-wide association study; polygenic risk score; protein-protein interaction; psoriasis; risk prediction.
Copyright © 2024 Antonatos, Koskeridis, Ralliou, Evangelou, Grafanaki, Georgiou, Tsilidis, Tzoulaki and Vasilopoulos.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genome-Wide Meta-analysis Identifies Risk Loci and Improves Disease Prediction of Age-Related Macular Degeneration.Ophthalmology. 2024 Jan;131(1):16-29. doi: 10.1016/j.ophtha.2023.08.023. Epub 2023 Aug 25. Ophthalmology. 2024. PMID: 37634759
-
A partitioned polygenic risk score reveals distinct contributions to psoriasis clinical phenotypes across a multi-ethnic cohort.J Transl Med. 2024 Sep 11;22(1):835. doi: 10.1186/s12967-024-05591-z. J Transl Med. 2024. PMID: 39261909 Free PMC article.
-
Polygenic risk scores for the prediction of common cancers in East Asians: A population-based prospective cohort study.Elife. 2023 Mar 27;12:e82608. doi: 10.7554/eLife.82608. Elife. 2023. PMID: 36971353 Free PMC article.
-
Polygenic risk scores: the future of cancer risk prediction, screening, and precision prevention.Med Rev (2021). 2022 Feb 14;1(2):129-149. doi: 10.1515/mr-2021-0025. eCollection 2021 Dec. Med Rev (2021). 2022. PMID: 37724297 Free PMC article. Review.
-
Genome-wide association studies and polygenic risk scores for skin cancer: clinically useful yet?Br J Dermatol. 2019 Dec;181(6):1146-1155. doi: 10.1111/bjd.17917. Epub 2019 Jul 7. Br J Dermatol. 2019. PMID: 30908599 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials

