Time-resolved single-cell transcriptomic sequencing
- PMID: 39568874
- PMCID: PMC11575584
- DOI: 10.1039/d4sc05700g
Time-resolved single-cell transcriptomic sequencing
Abstract
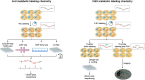
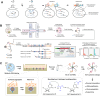
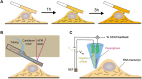
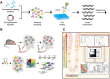
Cells experience continuous transformation under both physiological and pathological circumstances. Single-cell RNA sequencing (scRNA-seq) is competent in disclosing the disparities of cells; nevertheless, it poses challenges in linking the individual cell state at distinct time points. Although computational approaches based on scRNA-seq data have been put forward for trajectory analysis, the result is based on assumptions and fails to reflect the actual states. Consequently, it is necessary to incorporate a "time anchor" into the scRNA-seq library for the temporal documentation of the dynamic expression pattern. This review comprehensively overviews the time-resolved single-cell transcriptomic sequencing methodologies and applications. As scRNA-seq functions as the basis for profiling single-cell expression patterns, the review initially introduces various scRNA-seq approaches. Subsequently, the review focuses on the different experimental strategies for introducing a "time anchor" to scRNA-seq, highlighting their principles, strengths, weaknesses, and comparing their adaptation in various scenarios. Next, it provides a brief summary of applications in immunity response, cancer progression, and embryo development. Finally, the review concludes with a forward-looking perspective on future advancements in time-resolved single-cell transcriptomic sequencing.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures













References
-
- Erhard F. Saliba A.-E. Lusser A. Toussaint C. Hennig T. Prusty B. K. Kirschenbaum D. Abadie K. Miska E. A. Friedel C. C. Amit I. Micura R. Dölken L. Nat. Rev. Methods Primers. 2022;2:77. doi: 10.1038/s43586-022-00157-z. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

