Mechanical confinement matters: Unveiling the effect of two-photon polymerized 2.5D and 3D microarchitectures on neuronal YAP expression and neurite outgrowth
- PMID: 39569166
- PMCID: PMC11576396
- DOI: 10.1016/j.mtbio.2024.101325
Mechanical confinement matters: Unveiling the effect of two-photon polymerized 2.5D and 3D microarchitectures on neuronal YAP expression and neurite outgrowth
Abstract
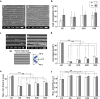
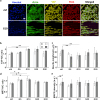
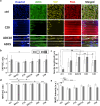
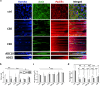
The effect of mechanical cues on cellular behaviour has been reported in multiple studies so far, and a specific aspect of interest is the role of mechanotransductive proteins in neuronal development. Among these, yes-associated protein (YAP) is responsible for multiple functions in neuronal development such as neuronal progenitor cells migration and differentiation while myocardin-related transcription factor A (MRTFA) facilitates neurite outgrowth and axonal pathfinding. Both proteins have indirectly intertwined fates via their signalling pathways. There is little literature investigating the roles of YAP and MRTFA in vitro concerning neurite outgrowth in mechanically confined microenvironments. Moreover, our understanding of their relationship in immature neurons cultured within engineered confined microenvironments is still lacking. In this study, we fabricated, via two-photon polymerization (2PP), 2.5D microgrooves and 3D polymeric microchannels, with a diameter range from 5 to 30 μm. We cultured SH-SY5Y cells and differentiated them into immature neuron-like cells on both 2.5D and 3D microstructures to investigate the effect of mechanical confinement on cell morphology and protein expression. In 2.5D microgrooves, both YAP and MRTFA nuclear/cytoplasmic (N/C) ratios exhibited maxima in the 10 μm grooves indicating a strong relation with mechanical-stress-inducing confinement. In 3D microchannels, both proteins' N/C ratio exhibited minima in presence of 5 or 10 μm channels, a behaviour that was opposite to the ones observed in the 2.5D microgrooves and that indicates how the geometry and mechanical confinement of 3D microenvironments are unique compared to 2.5D ones due to focal adhesion, actin, and nuclear polarization. Further, especially in presence of 2.5D microgrooves, cells featured an inversely proportional relationship between YAP N/C ratio and the average neurite length. Finally, we also cultured human induced pluripotent stem cells (hiPSCs) and differentiated them into cortical neurons on the microstructures for up to 2 weeks. Interestingly, YAP and MRTFA N/C ratios also showed a maximum around the 10 μm 2.5D microgrooves, indicating the physiological relevance of our study. Our results elucidate the possible differences induced by 2.5D and 3D confining microenvironments in neuronal development and paves the way for understanding the intricate interplay between mechanotransductive proteins and their effect on neural cell fate within engineered cell microenvironments.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures









Similar articles
-
cAMP response element-binding protein and Yes-associated protein form a feedback loop that promotes neurite outgrowth.J Cell Mol Med. 2018 Jan;22(1):374-381. doi: 10.1111/jcmm.13324. Epub 2017 Aug 31. J Cell Mol Med. 2018. PMID: 28857442 Free PMC article.
-
Micropatterning-retinoic acid co-control of neuronal cell morphology and neurite outgrowth.Acta Biomater. 2013 Jan;9(1):4592-8. doi: 10.1016/j.actbio.2012.08.039. Epub 2012 Aug 30. Acta Biomater. 2013. PMID: 22939924
-
Spatial confinement modulates endothelial cell behavior and traction force in 3D hydrogel microgrooves.Mater Today Bio. 2024 Apr 30;26:101074. doi: 10.1016/j.mtbio.2024.101074. eCollection 2024 Jun. Mater Today Bio. 2024. PMID: 38736613 Free PMC article.
-
Role of YAP as a Mechanosensing Molecule in Stem Cells and Stem Cell-Derived Hematopoietic Cells.Int J Mol Sci. 2022 Nov 23;23(23):14634. doi: 10.3390/ijms232314634. Int J Mol Sci. 2022. PMID: 36498961 Free PMC article. Review.
-
Control of cellular responses to mechanical cues through YAP/TAZ regulation.J Biol Chem. 2019 Nov 15;294(46):17693-17706. doi: 10.1074/jbc.REV119.007963. Epub 2019 Oct 8. J Biol Chem. 2019. PMID: 31594864 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

