Gene expression of tight junctions in foreskin is not affected by HIV pre-exposure prophylaxis
- PMID: 39569196
- PMCID: PMC11576434
- DOI: 10.3389/fimmu.2024.1415475
Gene expression of tight junctions in foreskin is not affected by HIV pre-exposure prophylaxis
Abstract
Introduction: Tight junctions (TJs) serve as permeability filters between the internal and external cellular environment. A large number of proteins have been identified to be localized at the TJs. Due to limitations in tissue collection, TJs in the male genital tract have been understudied.
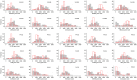
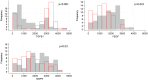
Methods: We analysed the transcriptomics of 132 TJ genes in foreskin tissue of men requesting voluntary medical male circumcision (VMMC) and enrolled in the Combined HIV Adolescent Prevention Study (CHAPS) trial conducted in South Africa and Uganda (NCT03986970). The trial evaluated the dose requirements for event-driven HIV pre-exposure prophylaxis (PrEP) with emtricitabine-tenofovir (FTC-TDF) or emtricitabine-tenofovir alafenamide (FTC-TAF) during insertive sex. A total of 144 participants were randomized to either control arm or one of 8 PrEP arms (n=16/arm), receiving oral FTC-TDF or FTC-TAF over one or two days. Following in vivo oral PrEP dosing and VMMC, the expression level of three important TJ proteins (CLDN-1, OCN and ZO-1) was measured ex vivo in foreskin tissue by Western blot. The expression of cytokine genes implicated in TJ regulation was determined. Non-parametric Kruskal-Wallis tests were used to compare TJ gene expression and protein levels by type of PrEP received, and Spearman's correlation coefficients were calculated to assess whether TJ gene expression levels were related to cytokine gene levels or to PrEP drug concentrations and their active intracellularly phosphorylated metabolites.
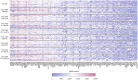
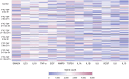
Results: A high level of expression in foreskin tissue was found for 118 (of 132) TJ genes analysed; this finding contributed to create a map of TJ components within the male genital tract. Importantly, PrEP regimens tested in the CHAPS trial did not affect the expression of TJ genes and the analysed proteins in the foreskin; thus, further supporting the safety of this prevention strategy against HIV-1 transmission during insertive sex. Additionally, we identified the level of several cytokines' genes to be correlated to TJ gene expression: among them, IL-18, IL-33 and VEGF.
Discussion: TJs can limit viral entry into target cells; to affect this biological function viruses can reduce the expression of TJ proteins. Our study, on the expression and regulation of TJs in the foreskin, contribute important knowledge for PrEP safety and further design of HIV-1 prophylaxis.
Keywords: cytokines; emtricitabine tenofovir; foreskin; pre-exposure prophylaxis PrEP; tight junctions; transcriptomes.
Copyright © 2024 Webb, Petkov, Yun, Else, Lebina, Serwanga, Pillay, Seiphetlo, Mugaba, Namubiru, Odoch, Opoka, Ssemata, Kaleebu, Khoo, Martinson, Fox, Gray, Herrera and Chiodi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

