G-quadruplex stabilizer CX-5461 effectively combines with radiotherapy to target α-thalassemia/mental retardation X-linked-deficient malignant glioma
- PMID: 39570009
- PMCID: PMC12083236
- DOI: 10.1093/neuonc/noae248
G-quadruplex stabilizer CX-5461 effectively combines with radiotherapy to target α-thalassemia/mental retardation X-linked-deficient malignant glioma
Abstract
Background: Inactivation of α-thalassemia/mental retardation X-linked (ATRX) represents a defining molecular feature in large subsets of malignant glioma. ATRX deficiency gives rise to abnormal G-quadruplex (G4) DNA secondary structures, enhancing replication stress and genomic instability. Building on earlier work, we evaluated the extent to which pharmacological G4 stabilization selectively enhances DNA damage and cell death in ATRX-deficient preclinical glioma models.
Methods: Using the G4 stabilizer CX-5461, we treated patient-derived glioma stem cells (GSCs) in vitro and GSC flank and intracranial murine xenografts in vivo to evaluate efficacy as both a single agent and in combination with ionizing radiation (IR), the latter a central element of current treatment standards.
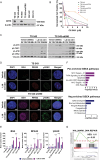
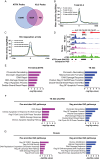
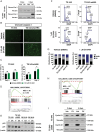
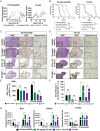
Results: CX-5461 promoted dose-sensitive lethality in ATRX-deficient GSCs relative to ATRX-intact controls. Mechanistic studies revealed that CX-5461 disrupted histone variant H3.3 deposition, enhanced replication stress and DNA damage, activated p53-independent apoptosis, and induced G2/M arrest to a greater extent in ATRX-deficient GSCs than in ATRX-intact counterparts. These data were corroborated in vivo, where CX-5461/IR treatment profoundly delayed tumor growth and prolonged survival in mice bearing ATRX-deficient flank xenografts. Histopathological analyses revealed decreased proliferation, increased apoptosis, and significant G4 induction, replication stress, and DNA damage in CX-5461-treated tumors, both alone and in combination with IR. Finally, despite suboptimal blood-brain-barrier penetration, systemic CX-5461 treatment induced tangible pharmacodynamic effects in ATRX-deficient intracranial GSC models.
Conclusions: In totality, our work substantively demonstrates efficacy and defines mechanisms of action for G4 stabilization as a novel therapeutic strategy targeting ATRX-deficient malignant glioma, laying the groundwork for clinical translation.
Keywords: ATRX; CX-5461; G-quadruplex; glioma; radiation.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
None declared.
Figures

References
-
- Levy MA, Kernohan KD, Jiang Y, Bérubé NG. ATRX promotes gene expression by facilitating transcriptional elongation through guanine-rich coding regions. Hum Mol Genet. 2015;24(7):1824–1835. - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA240338/CA/NCI NIH HHS/United States
- TL1 TR003169/TR/NCATS NIH HHS/United States
- UL1 TR003167/TR/NCATS NIH HHS/United States
- TL1TR003169 and UL1TR003167/National Center of Advancing Translational Sciences of the National Institutes of Health
- CA240338/National Institutes of Health/National Cancer Institute
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

