TIdeS: A Comprehensive Framework for Accurate Open Reading Frame Identification and Classification in Eukaryotic Transcriptomes
- PMID: 39570867
- PMCID: PMC11631190
- DOI: 10.1093/gbe/evae252
TIdeS: A Comprehensive Framework for Accurate Open Reading Frame Identification and Classification in Eukaryotic Transcriptomes
Abstract
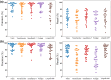
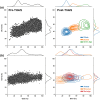
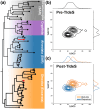
Studying fundamental aspects of eukaryotic biology through genetic information can face numerous challenges, including contamination and intricate biotic interactions, which are particularly pronounced when working with uncultured eukaryotes. However, existing tools for predicting open reading frames (ORFs) from transcriptomes are limited in these scenarios. Here we introduce Transcript Identification and Selection (TIdeS), a framework designed to address these nontrivial challenges associated with current 'omics approaches. Using transcriptomes from 32 taxa, representing the breadth of eukaryotic diversity, TIdeS outperforms most conventional ORF-prediction methods (i.e. TransDecoder), identifying a greater proportion of complete and in-frame ORFs. Additionally, TIdeS accurately classifies ORFs using minimal input data, even in the presence of "heavy contamination". This built-in flexibility extends to previously unexplored biological interactions, offering a robust single-stop solution for precise ORF predictions and subsequent decontamination. Beyond applications in phylogenomic-based studies, TIdeS provides a robust means to explore biotic interactions in eukaryotes (e.g. host-symbiont, prey-predator) and for reproducible dataset curation from transcriptomes and genomes.
Keywords: ORF prediction; biotic interactions; contamination; machine learning; phylogenomics.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Three-nucleotide periodicity of nucleotide diversity in a population enables the identification of open reading frames.Brief Bioinform. 2022 Jul 18;23(4):bbac210. doi: 10.1093/bib/bbac210. Brief Bioinform. 2022. PMID: 35698834 Free PMC article.
-
The random nature of genome architecture: predicting open reading frame distributions.PLoS One. 2009 Jul 30;4(7):e6456. doi: 10.1371/journal.pone.0006456. PLoS One. 2009. PMID: 19649247 Free PMC article.
-
Probing of plant transcriptomes reveals the hidden genetic diversity of the family Secoviridae.Arch Virol. 2024 Jun 20;169(7):150. doi: 10.1007/s00705-024-06076-6. Arch Virol. 2024. PMID: 38898334
-
The regulatory potential of upstream open reading frames in eukaryotic gene expression.Wiley Interdiscip Rev RNA. 2014 Nov-Dec;5(6):765-78. doi: 10.1002/wrna.1245. Epub 2014 Jul 3. Wiley Interdiscip Rev RNA. 2014. PMID: 24995549 Review.
-
Function and Evolution of Upstream ORFs in Eukaryotes.Trends Biochem Sci. 2019 Sep;44(9):782-794. doi: 10.1016/j.tibs.2019.03.002. Epub 2019 Apr 16. Trends Biochem Sci. 2019. PMID: 31003826 Review.
References
-
- Akiba T, Sano S, Yanase T, Ohta T, Koyama M. 2019. Optuna: a next-generation hyperparameter optimization framework. In: Proceedings of the 25th ACM SIGKDD international conference on knowledge discovery & data mining. New York, NY, USA: KDD ‘19 Association for Computing Machinery. pp. 2623–2631.
-
- Aponte A, Gyaltshen Y, Burns JA, Heiss AA, Kim E, Warring SD. The bacterial diversity lurking in protist cell cultures. Am Mus Novit. 2021:2021(3975):1–14. 10.1206/3975.1. - DOI
-
- Blaz J, Galindo LJ, Heiss AA, Kaur H, Torruella G, Yang A, Alexa Thompson L, Filbert A, Warring S, Narechania A, et al. One high quality genome and two transcriptome datasets for new species of Mantamonas, a deep-branching eukaryote clade. Sci Data. 2023:10(1):603. 10.1038/s41597-023-02488-2. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

