SARS-CoV-2 evolution balances conflicting roles of N protein phosphorylation
- PMID: 39571001
- PMCID: PMC11620656
- DOI: 10.1371/journal.ppat.1012741
SARS-CoV-2 evolution balances conflicting roles of N protein phosphorylation
Abstract
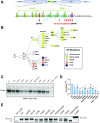
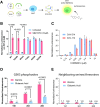
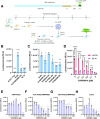
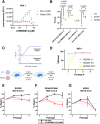
All lineages of SARS-CoV-2, the coronavirus responsible for the COVID-19 pandemic, contain mutations between amino acids 199 and 205 in the nucleocapsid (N) protein that are associated with increased infectivity. The effects of these mutations have been difficult to determine because N protein contributes to both viral replication and viral particle assembly during infection. Here, we used single-cycle infection and virus-like particle assays to show that N protein phosphorylation has opposing effects on viral assembly and genome replication. Ancestral SARS-CoV-2 N protein is densely phosphorylated, leading to higher levels of genome replication but 10-fold lower particle assembly compared to evolved variants with low N protein phosphorylation, such as Delta (N:R203M), Iota (N:S202R), and B.1.2 (N:P199L). A new open reading frame encoding a truncated N protein called N*, which occurs in the B.1.1 lineage and subsequent lineages of the Alpha, Gamma, and Omicron variants, supports high levels of both assembly and replication. Our findings help explain the enhanced fitness of viral variants of concern and a potential avenue for continued viral selection.
Copyright: © 2024 Syed et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
AMS and JAD are co-inventors on a patent application filed by Gladstone Institutes and University of California on the generation of SARS-CoV-2 VLPs. TYT and MO are inventors on a patent application filed by the Gladstone Institutes that covers the use of pGLUE to generate SARS-CoV-2 infectious clones and replicons. The NJK Laboratory has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. NJK has a financially compensated consulting agreement with Maze Therapeutics. NJK is the President and is on the Board of Directors of Rezo Therapeutics, and he is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, GEn1E Lifesciences, and Interline Therapeutics. All other authors declare no competing interests.
Figures
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

