Comparing microbiological and molecular diagnostic tools for the surveillance of anthrax
- PMID: 39571005
- PMCID: PMC11620650
- DOI: 10.1371/journal.pntd.0012122
Comparing microbiological and molecular diagnostic tools for the surveillance of anthrax
Abstract
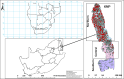
The diagnosis of anthrax, a zoonotic disease caused by Bacillus anthracis can be complicated by detection of closely related species. Conventional diagnosis of anthrax involves microscopy, culture identification of bacterial colonies and molecular detection. Genetic markers used are often virulence gene targets such as B. anthracis protective antigen (pagA, also called BAPA, occurring on plasmid pXO1), lethal factor (lef, on pXO1), capsule-encoding capB/C (located on pXO2) as well as chromosomal Ba-1. Combinations of genetic markers using real-time/quantitative polymerase chain reaction (qPCR) are used to confirm B. anthracis from culture but can also be used directly on diagnostic samples to avoid propagation and its associated biorisks and for faster identification. We investigated how the presence of closely related species could complicate anthrax diagnoses with and without culture to standardise the use of genetic markers using qPCR for accurate anthrax diagnosis. Using blood smears from 2012-2020 from wildlife mortalities (n = 1708) in Kruger National Park in South Africa where anthrax is endemic, we contrasted anthrax diagnostic results based on qPCR, microscopy, and culture. From smears, 113/1708 grew bacteria in culture, from which 506 isolates were obtained. Of these isolates, only 24.7% (125 isolates) were positive for B. anthracis based on genetic markers or microscopy. However, among these, merely 4/125 (3.2%) were confirmed B. anthracis isolates (based on morphology, microscopy, and sensitivity testing to penicillin and gamma-phage) from the blood smear, likely due to poor survival of spores on stored smears. This study identified B. cereus sensu lato, which included B. cereus and B. anthracis, Peribacillus spp., and Priestia spp. clusters using gyrB gene in selected bacterial isolates positive for pagA region using BAPA probe. Using qPCR on blood smears, 52.1% (890 samples) tested positive for B. anthracis based on one or a combination of genetic markers which included the 25 positive controls. Notably, the standard lef primer set displayed the lowest specificity and accuracy. The Ba-1+BAPA+lef combination showed 100% specificity, sensitivity, and accuracy. Various marker combinations, such as Ba-1+capB, BAPA+capB, Ba-1+BAPA+capB+lef, and BAPA+lef+capB, all demonstrated 100.0% specificity and 98.7% accuracy, while maintaining a sensitivity of 96.6%. Using Ba-1+BAPA+lef+capB, as well as Ba-1+BAPA+lef with molecular diagnosis accurately detects B. anthracis in the absence of bacterial culture. Systematically combining microscopy and molecular markers holds promise for notably reducing false positives. This significantly enhances the detection and surveillance of diseases like anthrax in southern Africa and beyond and reduces the need for propagation of the bacteria in culture.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A genome-based investigation of the Priestia species isolated from anthrax endemic regions in Kruger National Park.Infect Genet Evol. 2024 Sep;123:105649. doi: 10.1016/j.meegid.2024.105649. Epub 2024 Jul 25. Infect Genet Evol. 2024. PMID: 39059732
-
Polyphasic characterization of Bacillus species from anthrax outbreaks in animals from South Africa and Lesotho.J Infect Dev Ctries. 2016 Aug 31;10(8):814-23. doi: 10.3855/jidc.7798. J Infect Dev Ctries. 2016. PMID: 27580326
-
Bacillus anthracis genetics and virulence gene regulation.Curr Top Microbiol Immunol. 2002;271:143-64. doi: 10.1007/978-3-662-05767-4_7. Curr Top Microbiol Immunol. 2002. PMID: 12224521 Review.
-
In silico and in vitro evaluation of PCR-based assays for the detection of Bacillus anthracis chromosomal signature sequences.Virulence. 2013 Nov 15;4(8):671-85. doi: 10.4161/viru.26288. Epub 2013 Sep 9. Virulence. 2013. PMID: 24005110 Free PMC article.
-
Bacillus anthracis physiology and genetics.Mol Aspects Med. 2009 Dec;30(6):386-96. doi: 10.1016/j.mam.2009.07.004. Epub 2009 Aug 3. Mol Aspects Med. 2009. PMID: 19654018 Free PMC article. Review.
Cited by
-
Decoding the anomalies: a genome-based analysis of Bacillus cereus group strains closely related to Bacillus anthracis.Front Microbiol. 2025 Feb 5;16:1527049. doi: 10.3389/fmicb.2025.1527049. eCollection 2025. Front Microbiol. 2025. PMID: 39973926 Free PMC article.
References
-
- Ben-Noun L. [Characteristics of anthrax: its description and biblical name—Shehin]. Harefuah. 2002;141 Spec No:4–6, 124. - PubMed
-
- W.H.O. Anthrax in humans and animals. In: Organization WH, editor. World Health Organization. 4. 4 ed. World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland: WHO Press; 2008. p. 1–33.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

